Association between resting-state functional brain connectivity and gene expression is altered in autism spectrum disorder
- PMID: 35680911
- PMCID: PMC9184501
- DOI: 10.1038/s41467-022-31053-5
Association between resting-state functional brain connectivity and gene expression is altered in autism spectrum disorder
Abstract
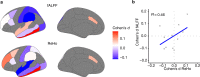
Gene expression covaries with brain activity as measured by resting state functional magnetic resonance imaging (MRI). However, it is unclear how genomic differences driven by disease state can affect this relationship. Here, we integrate from the ABIDE I and II imaging cohorts with datasets of gene expression in brains of neurotypical individuals and individuals with autism spectrum disorder (ASD) with regionally matched brain activity measurements from fMRI datasets. We identify genes linked with brain activity whose association is disrupted in ASD. We identified a subset of genes that showed a differential developmental trajectory in individuals with ASD compared with controls. These genes are enriched in voltage-gated ion channels and inhibitory neurons, pointing to excitation-inhibition imbalance in ASD. We further assessed differences at the regional level showing that the primary visual cortex is the most affected region in ASD. Our results link disrupted brain expression patterns of individuals with ASD to brain activity and show developmental, cell type, and regional enrichment of activity linked genes.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Grasby, K. L. et al. The genetic architecture of the human cerebral cortex. Science10.1126/science.aay6690 (2020).
Publication types
MeSH terms
Grants and funding
- R01 MH094714/MH/NIMH NIH HHS/United States
- P50 MH106438/MH/NIMH NIH HHS/United States
- R21 NS106447/NS/NINDS NIH HHS/United States
- P50 HD103557/HD/NICHD NIH HHS/United States
- R01 MH110927/MH/NIMH NIH HHS/United States
- R01 MH121521/MH/NIMH NIH HHS/United States
- U01 MH115746/MH/NIMH NIH HHS/United States
- R01 MH126481/MH/NIMH NIH HHS/United States
- R01 MH102603/MH/NIMH NIH HHS/United States
- R01 MH123922/MH/NIMH NIH HHS/United States
- R01 MH109912/MH/NIMH NIH HHS/United States
- UF1 NS115821/NS/NINDS NIH HHS/United States
- R01 HG011641/HG/NHGRI NIH HHS/United States

