Extracellular release of two peptidases dominates generation of the trypanosome quorum-sensing signal
- PMID: 35680928
- PMCID: PMC9184580
- DOI: 10.1038/s41467-022-31057-1
Extracellular release of two peptidases dominates generation of the trypanosome quorum-sensing signal
Abstract
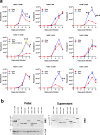
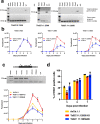
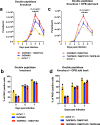
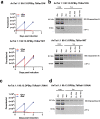
Trypanosomes causing African sleeping sickness use quorum-sensing (QS) to generate transmission-competent stumpy forms in mammalian hosts. This density-dependent process is signalled by oligopeptides that stimulate the signal transduction pathway leading to stumpy formation. Here, using mass spectrometry analysis, we identify peptidases released by trypanosomes and, for 12 peptidases, confirm their extracellular delivery. Thereafter, we determine the contribution of each peptidase to QS signal production using systematic inducible overexpression in vivo, and confirm this activity operates through the physiological QS signalling pathway. Gene knockout of the QS-active peptidases identifies two enzymes, oligopeptidase B and metallocarboxypeptidase 1, that significantly reduce QS when ablated individually. Further, combinatorial gene knockout of both peptidases confirms their dominance in the generation of the QS signal, with peptidase release of oligopeptidase B mediated via an unconventional protein secretion pathway. This work identifies how the QS signal driving trypanosome virulence and transmission is generated in mammalian hosts.
© 2022. The Author(s).
Conflict of interest statement
We declare that the authors have no competing interests as defined by Nature Portfolio, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Figures



References
-
- Buscher, P., Cecchi, G., Jamonneau, V. & Priotto, G. Human African trypanosomiasis. Lancet390, 2397–2409 (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

