Genomic analysis defines clonal relationships of ductal carcinoma in situ and recurrent invasive breast cancer
- PMID: 35681052
- PMCID: PMC9197769
- DOI: 10.1038/s41588-022-01082-3
Genomic analysis defines clonal relationships of ductal carcinoma in situ and recurrent invasive breast cancer
Abstract
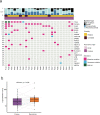
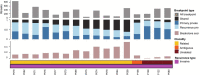
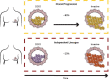
Ductal carcinoma in situ (DCIS) is the most common form of preinvasive breast cancer and, despite treatment, a small fraction (5-10%) of DCIS patients develop subsequent invasive disease. A fundamental biologic question is whether the invasive disease arises from tumor cells in the initial DCIS or represents new unrelated disease. To address this question, we performed genomic analyses on the initial DCIS lesion and paired invasive recurrent tumors in 95 patients together with single-cell DNA sequencing in a subset of cases. Our data show that in 75% of cases the invasive recurrence was clonally related to the initial DCIS, suggesting that tumor cells were not eliminated during the initial treatment. Surprisingly, however, 18% were clonally unrelated to the DCIS, representing new independent lineages and 7% of cases were ambiguous. This knowledge is essential for accurate risk evaluation of DCIS, treatment de-escalation strategies and the identification of predictive biomarkers.
© 2022. The Author(s).
Conflict of interest statement
H.R.D. and S.N.Z. hold patent filings on algorithms for tumor classification (PCT/EP2017/060294PCT/EP2017/060289, PCT/EP2017/060279). The remaining authors declare no competing interests.
Figures










References
Publication types
MeSH terms
Substances
Grants and funding
- 28990/CRUK_/Cancer Research UK/United Kingdom
- R01 CA185138/CA/NCI NIH HHS/United States
- C60100/A23916/CRUK_/Cancer Research UK/United Kingdom
- R01 CA240526/CA/NCI NIH HHS/United States
- A24043/CRUK_/Cancer Research UK/United Kingdom
- BRC-125-20014/DH_/Department of Health/United Kingdom
- P20 GM130423/GM/NIGMS NIH HHS/United States
- P30 CA014236/CA/NCI NIH HHS/United States
- C17422/A25154/CRUK_/Cancer Research UK/United Kingdom
- 2014NOVTAP379/BBC_/Breast Cancer Now/United Kingdom
- P30 CA016672/CA/NCI NIH HHS/United States
- 23916/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Medical

