In vivo engineered B cells secrete high titers of broadly neutralizing anti-HIV antibodies in mice
- PMID: 35681059
- PMCID: PMC7613293
- DOI: 10.1038/s41587-022-01328-9
In vivo engineered B cells secrete high titers of broadly neutralizing anti-HIV antibodies in mice
Abstract
Transplantation of B cells engineered ex vivo to secrete broadly neutralizing antibodies (bNAbs) has shown efficacy in disease models. However, clinical translation of this approach would require specialized medical centers, technically demanding protocols and major histocompatibility complex compatibility of donor cells and recipients. Here we report in vivo B cell engineering using two adeno-associated viral vectors, with one coding for Staphylococcus aureus Cas9 (saCas9) and the other for 3BNC117, an anti-HIV bNAb. After intravenously injecting the vectors into mice, we observe successful editing of B cells leading to memory retention and bNAb secretion at neutralizing titers of up to 6.8 µg ml-1. We observed minimal clustered regularly interspaced palindromic repeats (CRISPR)-Cas9 off-target cleavage as detected by unbiased CHANGE-sequencing analysis, whereas on-target cleavage in undesired tissues is reduced by expressing saCas9 from a B cell-specific promoter. In vivo B cell engineering to express therapeutic antibodies is a safe, potent and scalable method, which may be applicable not only to infectious diseases but also in the treatment of noncommunicable conditions, such as cancer and autoimmune disease.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures





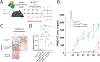




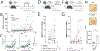
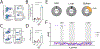



Comment in
-
CRISPR comes a-knock-in to reprogram antibodies in vivo.Nat Biotechnol. 2022 Aug;40(8):1183-1184. doi: 10.1038/s41587-022-01299-x. Nat Biotechnol. 2022. PMID: 35681058 No abstract available.
-
Long-term control of HIV.Nat Rev Microbiol. 2022 Aug;20(8):446. doi: 10.1038/s41579-022-00759-0. Nat Rev Microbiol. 2022. PMID: 35705741 No abstract available.
-
B cell engineering in vivo: Accelerating induction of broadly neutralizing antibodies against HIV-1 infection.Signal Transduct Target Ther. 2023 Jan 6;8(1):13. doi: 10.1038/s41392-022-01269-4. Signal Transduct Target Ther. 2023. PMID: 36604417 Free PMC article. No abstract available.
References
METHODS REFERENCES
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

