Limited nutrient availability in the tumor microenvironment renders pancreatic tumors sensitive to allosteric IDH1 inhibitors
- PMID: 35681100
- PMCID: PMC9325670
- DOI: 10.1038/s43018-022-00393-y
Limited nutrient availability in the tumor microenvironment renders pancreatic tumors sensitive to allosteric IDH1 inhibitors
Abstract
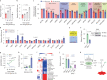
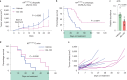
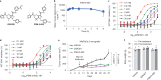
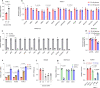
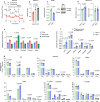
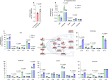
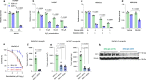
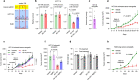
Nutrient-deprived conditions in the tumor microenvironment (TME) restrain cancer cell viability due to increased free radicals and reduced energy production. In pancreatic cancer cells a cytosolic metabolic enzyme, wild-type isocitrate dehydrogenase 1 (wtIDH1), enables adaptation to these conditions. Under nutrient starvation, wtIDH1 oxidizes isocitrate to generate α-ketoglutarate (αKG) for anaplerosis and NADPH to support antioxidant defense. In this study, we show that allosteric inhibitors of mutant IDH1 (mIDH1) are potent wtIDH1 inhibitors under conditions present in the TME. We demonstrate that low magnesium levels facilitate allosteric inhibition of wtIDH1, which is lethal to cancer cells when nutrients are limited. Furthermore, the Food & Drug Administration (FDA)-approved mIDH1 inhibitor ivosidenib (AG-120) dramatically inhibited tumor growth in preclinical models of pancreatic cancer, highlighting this approach as a potential therapeutic strategy against wild-type IDH1 cancers.
© 2022. This is a U.S. government work and not under copyright protection in the U.S.; foreign copyright protection may apply.
Conflict of interest statement
R.G. is an inventor on multiple issued patents and patent applications covering the manufacture and use of 3DNA technology as part of his role in Code Biotherapeutics, Inc. He is also an employee and officer of Code Biotherapeutics, Inc. and has stock and stock options. C.A.L. has received consulting fees from Astellas Pharmaceuticals and Odyssey Therapeutics and is an inventor on patents pertaining to Kras-regulated metabolic pathways, redox control pathways in cancer and targeting of the GOT1-pathway as a therapeutic approach. J.M.S. is a coauthor on patents on IDH1 inhibitors, and has received sponsored research funding from the Barer Institute and patents pending to Wistar Institute. J.M.W., along with University Hospitals, filed the following patent application on 24 September 2020: Methods for Treating Wild Type Isocitrate Dehydrogenase 1 Cancers. Information regarding this patent application is as follows: PCT/US20/52445 filed 24 September 2020, Claiming Priority to US 62/911,717 filed 7 October 2019, file nos: UHOSP-19738 | 2019-014.
Figures








Similar articles
-
Wild-type IDH1 inhibition enhances chemotherapy response in melanoma.J Exp Clin Cancer Res. 2022 Sep 24;41(1):283. doi: 10.1186/s13046-022-02489-w. J Exp Clin Cancer Res. 2022. PMID: 36153582 Free PMC article.
-
Targeting wild-type IDH1 enhances chemosensitivity in pancreatic cancer.bioRxiv [Preprint]. 2023 Mar 29:2023.03.29.534596. doi: 10.1101/2023.03.29.534596. bioRxiv. 2023. Update in: Cancer Res. 2024 Sep 16;84(18):3072-3085. doi: 10.1158/0008-5472.CAN-23-1895. PMID: 37034685 Free PMC article. Updated. Preprint.
-
Preclinical Drug Metabolism, Pharmacokinetic, and Pharmacodynamic Profiles of Ivosidenib, an Inhibitor of Mutant Isocitrate Dehydrogenase 1 for Treatment of Isocitrate Dehydrogenase 1-Mutant Malignancies.Drug Metab Dispos. 2021 Oct;49(10):870-881. doi: 10.1124/dmd.120.000234. Epub 2021 Jul 28. Drug Metab Dispos. 2021. PMID: 34321251
-
Looking Beyond the Surface: Olutasidenib and Ivosidenib for Treatment of mIDH1 Acute Myeloid Leukemia.Curr Treat Options Oncol. 2024 Nov;25(11):1345-1353. doi: 10.1007/s11864-024-01264-7. Epub 2024 Oct 16. Curr Treat Options Oncol. 2024. PMID: 39406957 Free PMC article. Review.
-
Inhibitors of Mutant Isocitrate Dehydrogenases 1 and 2 (mIDH1/2): An Update and Perspective.J Med Chem. 2018 Oct 25;61(20):8981-9003. doi: 10.1021/acs.jmedchem.8b00159. Epub 2018 Jun 18. J Med Chem. 2018. PMID: 29847930 Review.
Cited by
-
The pentose phosphate pathway in health and disease.Nat Metab. 2023 Aug;5(8):1275-1289. doi: 10.1038/s42255-023-00863-2. Epub 2023 Aug 23. Nat Metab. 2023. PMID: 37612403 Free PMC article. Review.
-
Glutamine addiction and therapeutic strategies in pancreatic cancer.World J Gastrointest Oncol. 2023 Nov 15;15(11):1852-1863. doi: 10.4251/wjgo.v15.i11.1852. World J Gastrointest Oncol. 2023. PMID: 38077649 Free PMC article. Review.
-
Use of Three-Dimensional Cell Culture Models in Drug Assays for Anti-Cancer Agents in Oral Cancer: Protocol for a Scoping Review.J Pers Med. 2023 Nov 17;13(11):1618. doi: 10.3390/jpm13111618. J Pers Med. 2023. PMID: 38003933 Free PMC article.
-
Patient-Derived Models of Cancer in the NCI PDMC Consortium: Selection, Pitfalls, and Practical Recommendations.Cancers (Basel). 2024 Jan 29;16(3):565. doi: 10.3390/cancers16030565. Cancers (Basel). 2024. PMID: 38339316 Free PMC article. Review.
-
Mapping Cell-in-Cell Structures in Oral Squamous Cell Carcinoma.Cells. 2023 Oct 8;12(19):2418. doi: 10.3390/cells12192418. Cells. 2023. PMID: 37830632 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- S10 OD030245/OD/NIH HHS/United States
- R01 CA196643/CA/NCI NIH HHS/United States
- P30 CA010815/CA/NCI NIH HHS/United States
- F32 CA247466/CA/NCI NIH HHS/United States
- P50 CA150964/CA/NCI NIH HHS/United States
- P30 CA043703/CA/NCI NIH HHS/United States
- R37 CA227865/CA/NCI NIH HHS/United States
- R33 DK070291/DK/NIDDK NIH HHS/United States
- P30 CA046592/CA/NCI NIH HHS/United States
- R01 CA248160/CA/NCI NIH HHS/United States
- R37 CA237421/CA/NCI NIH HHS/United States
- R01 CA212600/CA/NCI NIH HHS/United States
- R01 CA244931/CA/NCI NIH HHS/United States
- 29996/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

