Genome-wide identification and expression analysis of the R2R3-MYB gene family in tobacco (Nicotiana tabacum L.)
- PMID: 35681121
- PMCID: PMC9178890
- DOI: 10.1186/s12864-022-08658-7
Genome-wide identification and expression analysis of the R2R3-MYB gene family in tobacco (Nicotiana tabacum L.)
Abstract
Background: The R2R3-MYB transcription factor is one of the largest gene families in plants and involved in the regulation of plant development, hormone signal transduction, biotic and abiotic stresses. Tobacco is one of the most important model plants. Therefore, it will be of great significance to investigate the R2R3-MYB gene family and their expression patterns under abiotic stress and senescence in tobacco.
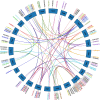
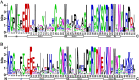
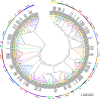
Results: A total of 174 R2R3-MYB genes were identified from tobacco (Nicotiana tabacum L.) genome and were divided into 24 subgroups based on phylogenetic analysis. Gene structure (exon/intron) and protein motifs were especially conserved among the NtR2R3-MYB genes, especially members within the same subgroup. The NtR2R3-MYB genes were distributed on 24 tobacco chromosomes. Analysis of gene duplication events obtained 3 pairs of tandem duplication genes and 62 pairs of segmental duplication genes, suggesting that segmental duplications is the major pattern for R2R3-MYB gene family expansion in tobacco. Cis-regulatory elements of the NtR2R3-MYB promoters were involved in cellular development, phytohormones, environmental stress and photoresponsive. Expression profile analysis showed that NtR2R3-MYB genes were widely expressed in different maturity tobacco leaves, and however, the expression patterns of different members appeared to be diverse. The qRT-PCR analysis of 15 NtR2R3-MYBs confirmed their differential expression under different abiotic stresses (cold, salt and drought), and notably, NtMYB46 was significantly up-regulated under three treatments.
Conclusions: In summary, a genome-wide identification, evolutionary and expression analysis of R2R3-MYB gene family in tobacco were conducted. Our results provided a solid foundation for further biological functional study of NtR2R3-MYB genes in tobacco.
Keywords: Gene expression; Nicotiana tobacum L.; Phylogenetic analysis; R2R3-MYB transcription factors; Stress response.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987;6(12):3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x. - DOI - PMC - PubMed
-
- Ogata K, Kanei-Ishii C, Sasaki M, Hatanaka H, Nagadoi A, Enari M, Nakamura H, Nishimura Y, Ishii S, Sarai A. The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat Struct Biol. 1996;3(2):178–187. doi: 10.1038/nsb0296-178. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

