Chemometric Differentiation of Sole and Plaice Fish Fillets Using Three Near-Infrared Instruments
- PMID: 35681393
- PMCID: PMC9180159
- DOI: 10.3390/foods11111643
Chemometric Differentiation of Sole and Plaice Fish Fillets Using Three Near-Infrared Instruments
Abstract
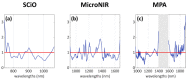
Fish species substitution is one of the most common forms of fraud all over the world, as fish identification can be very challenging for both consumers and experienced inspectors in the case of fish sold as fillets. The difficulties in distinguishing among different species may generate a "grey area" in which mislabelling can occur. Thus, the development of fast and reliable tools able to detect such frauds in the field is of crucial importance. In this study, we focused on the distinction between two flatfish species largely available on the market, namely the Guinean sole (Synaptura cadenati) and European plaice (Pleuronectes platessa), which are very similar looking. Fifty fillets of each species were analysed using three near-infrared (NIR) instruments: the handheld SCiO (Consumer Physics), the portable MicroNIR (VIAVI), and the benchtop MPA (Bruker). PLS-DA classification models were built using the spectral datasets, and all three instruments provided very good results, showing high accuracy: 94.1% for the SCiO and MicroNIR portable instruments, and 90.1% for the MPA benchtop spectrometer. The good classification results of the approach combining NIR spectroscopy, and simple chemometric classification methods suggest great applicability directly in the context of real-world marketplaces, as well as in official control plans.
Keywords: European plaice; Guinean sole; NIR; chemometrics; fish fillets; food fraud.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Reilly A. Overview of Food Fraud in the Fisheries Sector. FAO; Rome, Italy: 2018.
-
- Calosso M.C., Claydon J.A.B., Mariani S., Cawthorn D.M. Global footprint of mislabelled seafood on a small island nation. Biol. Conserv. 2020;245:108557. doi: 10.1016/j.biocon.2020.108557. - DOI
-
- Donlan C.J., Luque G.M. Exploring the causes of seafood fraud: A meta-analysis on mislabeling and price. Mar. Policy. 2019;100:258–264. doi: 10.1016/j.marpol.2018.11.022. - DOI
-
- Carvalho D.C., Guedes D., da Gloria Trindade M., Coelho R.M.S., de Lima Araujo P.H. Nationwide Brazilian governmental forensic programme reveals seafood mislabelling trends and rates using DNA barcoding. Fish. Res. 2017;191:30–35. doi: 10.1016/j.fishres.2017.02.021. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

