Role of Cytoskeletal Diaphanous-Related Formins in Hearing Loss
- PMID: 35681420
- PMCID: PMC9179844
- DOI: 10.3390/cells11111726
Role of Cytoskeletal Diaphanous-Related Formins in Hearing Loss
Abstract
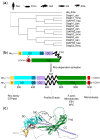
Hearing relies on the proper functioning of auditory hair cells and on actin-based cytoskeletal structures. Diaphanous-related formins (DRFs) are evolutionarily conserved cytoskeletal proteins that regulate the nucleation of linear unbranched actin filaments. They play key roles during metazoan development, and they seem particularly pivotal for the correct physiology of the reproductive and auditory systems. Indeed, in Drosophila melanogaster, a single diaphanous (dia) gene is present, and mutants show sterility and impaired response to sound. Vertebrates, instead, have three orthologs of the diaphanous gene: DIAPH1, DIAPH2, and DIAPH3. In humans, defects in DIAPH1 and DIAPH3 have been associated with different types of hearing loss. In particular, heterozygous mutations in DIAPH1 are responsible for autosomal dominant deafness with or without thrombocytopenia (DFNA1, MIM #124900), whereas regulatory mutations inducing the overexpression of DIAPH3 cause autosomal dominant auditory neuropathy 1 (AUNA1, MIM #609129). Here, we provide an overview of the expression and function of DRFs in normal hearing and deafness.
Keywords: DIAPH1; DIAPH2; DIAPH3; autosomal dominant auditory neuropathy; diaphanous-related formins; inherited sensorineural hearing loss; mouse mutants.
Conflict of interest statement
The authors declare no conflict of interest.
Figures