The Rice Serine/Arginine Splicing Factor RS33 Regulates Pre-mRNA Splicing during Abiotic Stress Responses
- PMID: 35681491
- PMCID: PMC9180459
- DOI: 10.3390/cells11111796
The Rice Serine/Arginine Splicing Factor RS33 Regulates Pre-mRNA Splicing during Abiotic Stress Responses
Abstract
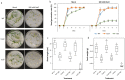
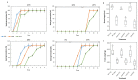
Abiotic stresses profoundly affect plant growth and development and limit crop productivity. Pre-mRNA splicing is a major form of gene regulation that helps plants cope with various stresses. Serine/arginine (SR)-rich splicing factors play a key role in pre-mRNA splicing to regulate different biological processes under stress conditions. Alternative splicing (AS) of SR transcripts and other transcripts of stress-responsive genes generates multiple splice isoforms that contribute to protein diversity, modulate gene expression, and affect plant stress tolerance. Here, we investigated the function of the plant-specific SR protein RS33 in regulating pre-mRNA splicing and abiotic stress responses in rice. The loss-of-function mutant rs33 showed increased sensitivity to salt and low-temperature stresses. Genome-wide analyses of gene expression and splicing in wild-type and rs33 seedlings subjected to these stresses identified multiple splice isoforms of stress-responsive genes whose AS are regulated by RS33. The number of RS33-regulated genes was much higher under low-temperature stress than under salt stress. Our results suggest that the plant-specific splicing factor RS33 plays a crucial role during plant responses to abiotic stresses.
Keywords: SR proteins; abiotic stress; alternative splicing; genome engineering; pre-mRNA splicing.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
Figures



References
-
- Chechanovsky N., Hovav R., Frenkel R., Faigenboim A., Eselson Y., Petreikov M., Moy M., Shen S., Schaffer A.A. Low temperature upregulates cwp expression and modifies alternative splicing patterns, increasing the severity of cwp-induced tomato fruit cuticular microfissures. Hortic. Res. 2019;6:122. doi: 10.1038/s41438-019-0204-9. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

