Proteomic Analysis of Lung Cancer Types-A Pilot Study
- PMID: 35681609
- PMCID: PMC9179298
- DOI: 10.3390/cancers14112629
Proteomic Analysis of Lung Cancer Types-A Pilot Study
Abstract
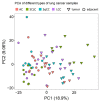
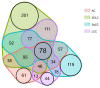
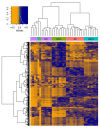
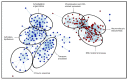
Lung cancer is the leading cause of tumor-related mortality, therefore significant effort is directed towards understanding molecular alterations occurring at the origin of the disease to improve current treatment options. The aim of our pilot-scale study was to carry out a detailed proteomic analysis of formalin-fixed paraffin-embedded tissue sections from patients with small cell or non-small cell lung cancer (adenocarcinoma, squamous cell carcinoma, and large cell carcinoma). Tissue surface digestion was performed on relatively small cancerous and tumor-adjacent normal regions and differentially expressed proteins were identified using label-free quantitative mass spectrometry and subsequent statistical analysis. Principal component analysis clearly distinguished cancerous and cancer adjacent normal samples, while the four lung cancer types investigated had distinct molecular profiles and gene set enrichment analysis revealed specific dysregulated biological processes as well. Furthermore, proteins with altered expression unique to a specific lung cancer type were identified and could be the targets of future studies.
Keywords: NSCLC; SCLC; cancer; gene set enrichment analysis; human tissue; lung cancer; lung tissue; mass spectrometry; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Pender A., Popat S. Understanding lung cancer molecular subtypes. Clin. Pract. 2014;11:441–453. doi: 10.2217/cpr.14.39. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

