Targeting Immunosuppressive Tumor-Associated Macrophages Using Innate T Cells for Enhanced Antitumor Reactivity
- PMID: 35681730
- PMCID: PMC9179365
- DOI: 10.3390/cancers14112749
Targeting Immunosuppressive Tumor-Associated Macrophages Using Innate T Cells for Enhanced Antitumor Reactivity
Abstract
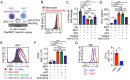
The field of T cell-based and chimeric antigen receptor (CAR)-engineered T (CAR-T) cell-based antitumor immunotherapy has seen substantial developments in the past decade; however, considerable issues, such as graft-versus-host disease (GvHD) and tumor-associated immunosuppression, have proven to be substantial roadblocks to widespread adoption and implementation. Recent developments in innate immune cell-based CAR therapy have opened several doors for the expansion of this therapy, especially as it relates to allogeneic cell sources and solid tumor infiltration. This study establishes in vitro killing assays to examine the TAM-targeting efficacy of MAIT, iNKT, and γδT cells. This study also assesses the antitumor ability of CAR-engineered innate T cells, evaluating their potential adoption for clinical therapies. The in vitro trials presented in this study demonstrate the considerable TAM-killing abilities of all three innate T cell types, and confirm the enhanced antitumor abilities of CAR-engineered innate T cells. The tumor- and TAM-targeting capacity of these innate T cells suggest their potential for antitumor therapy that supplements cytotoxicity with remediation of tumor microenvironment (TME)-immunosuppression.
Keywords: cancer immunotherapy; gamma delta T (γδT) cells; innate T cell; invariant natural killer T (iNKT) cell; mucosal-associated invariant T (MAIT) cell; tumor microenvironment (TME); tumor-associated macrophage.
Conflict of interest statement
L.Y. is a scientific advisor to AlzChem and Amberstone Biosciences, and a co-founder, stockholder, and advisory board member of Appia Bio. None of the declared companies contributed to or directed any of the research reported in this article. The present authors declare no competing interests.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

