LncRNA ZNF582-AS1 Expression and Methylation in Breast Cancer and Its Biological and Clinical Implications
- PMID: 35681777
- PMCID: PMC9179509
- DOI: 10.3390/cancers14112788
LncRNA ZNF582-AS1 Expression and Methylation in Breast Cancer and Its Biological and Clinical Implications
Abstract
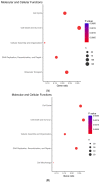
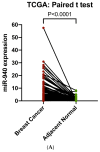
Background: Long non-coding RNAs (lncRNAs) play an important role in cellular activities and functions, but our understanding of their involvement in cancer is limited. Methods: TCGA data on RNA expression and DNA methylation were analyzed for lncRNAs' association with breast cancer survival, using the Cox proportional hazard regression model. Fresh tumor samples and clinical information from 361 breast cancer patients in our study were used to confirm the TCGA finding on ZNF582-AS1. A RT-qPCR method was developed to measure ZNF582-AS1 expression. Survival associations with ZNF582-AS1 were verified with a meta-analysis. In silico predictions of molecular targets and cellular functions of ZNF582-AS1 were performed based on its molecular signatures and nucleotide sequences. Results:ZNF582-AS1 expression was lower in breast tumors than adjacent normal tissues. Low ZNF582-AS1 was associated with high-grade or ER-negative tumors. Patients with high ZNF582-AS1 had a lower risk of relapse and death. These survival associations were confirmed in a meta-analysis and remained significant after adjustment for tumor grade, disease stage, patient age, and hormone receptor status. Correlation analysis indicated the possible suppression of ZNF582-AS1 expression by promoter methylation. Bioinformatics interrogation of molecular signatures suggested that ZNF582-AS1 could suppress tumor cell proliferation via downregulating the HER2-mediated signaling pathway. Analysis of online data also suggested that HIF-1-related transcription factors could suppress ZNF582-AS1 expression, and the lncRNA might bind to hsa-miR-940, a known oncogenic miRNA in breast cancer. Conclusions: ZNF582-AS1 may play a role in suppressing breast cancer progression. Elucidating the lncRNA's function and regulation may improve our understanding of the disease.
Keywords: ZNF582-AS1; breast cancer; lncRNA; methylation; prognosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Construction of a predictive model for breast cancer metastasis based on lncRNAs.Transl Cancer Res. 2023 Feb 28;12(2):387-397. doi: 10.21037/tcr-23-129. Epub 2023 Feb 27. Transl Cancer Res. 2023. PMID: 36915588 Free PMC article.
-
High methylation of the same promoter of lncRNA ZNF582-AS1/ZNF582 promotes malignant progression of esophageal cancer.Epigenomics. 2024;16(10):733-752. doi: 10.1080/17501911.2024.2342229. Epub 2024 Jun 4. Epigenomics. 2024. PMID: 38869483 Free PMC article.
-
Downregulation of lncRNA ZNF582-AS1 due to DNA hypermethylation promotes clear cell renal cell carcinoma growth and metastasis by regulating the N(6)-methyladenosine modification of MT-RNR1.J Exp Clin Cancer Res. 2021 Mar 10;40(1):92. doi: 10.1186/s13046-021-01889-8. J Exp Clin Cancer Res. 2021. Retraction in: J Exp Clin Cancer Res. 2023 Jun 6;42(1):144. doi: 10.1186/s13046-023-02719-9. PMID: 33691743 Free PMC article. Retracted.
-
A genomic screen for long noncoding RNA genes epigenetically silenced by aberrant DNA methylation in colorectal cancer.Sci Rep. 2016 May 24;6:26699. doi: 10.1038/srep26699. Sci Rep. 2016. PMID: 27215978 Free PMC article.
-
The role of long non-coding RNA AFAP1-AS1 in human malignant tumors.Pathol Res Pract. 2018 Oct;214(10):1524-1531. doi: 10.1016/j.prp.2018.08.014. Epub 2018 Aug 20. Pathol Res Pract. 2018. PMID: 30173945 Review.
Cited by
-
Construction of a predictive model for breast cancer metastasis based on lncRNAs.Transl Cancer Res. 2023 Feb 28;12(2):387-397. doi: 10.21037/tcr-23-129. Epub 2023 Feb 27. Transl Cancer Res. 2023. PMID: 36915588 Free PMC article.
-
High methylation of the same promoter of lncRNA ZNF582-AS1/ZNF582 promotes malignant progression of esophageal cancer.Epigenomics. 2024;16(10):733-752. doi: 10.1080/17501911.2024.2342229. Epub 2024 Jun 4. Epigenomics. 2024. PMID: 38869483 Free PMC article.
-
LINC00969 inhibits proliferation with metastasis of breast cancer by regulating phosphorylation of PI3K/AKT and ILP2 expression through HOXD8.PeerJ. 2023 Dec 18;11:e16679. doi: 10.7717/peerj.16679. eCollection 2023. PeerJ. 2023. PMID: 38130932 Free PMC article.
-
Long non-coding RNA AC099850.4 correlates with advanced disease state and predicts worse prognosis in triple-negative breast cancer.Front Med (Lausanne). 2023 Aug 31;10:1149860. doi: 10.3389/fmed.2023.1149860. eCollection 2023. Front Med (Lausanne). 2023. PMID: 37727755 Free PMC article.
-
Targeting the Vulnerabilities of Oncogene Activation.Cancers (Basel). 2023 Jun 27;15(13):3359. doi: 10.3390/cancers15133359. Cancers (Basel). 2023. PMID: 37444469 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

