Phenotypic and Genetic Characterization of Klebsiella pneumoniae Isolates from Wild Animals in Central Italy
- PMID: 35681810
- PMCID: PMC9179660
- DOI: 10.3390/ani12111347
Phenotypic and Genetic Characterization of Klebsiella pneumoniae Isolates from Wild Animals in Central Italy
Abstract
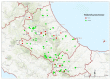
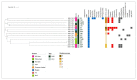
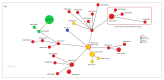
Despite Klebsiella pneumoniae being widely recognized as a nosocomial pathogen, there is a critical lack in defining its reservoirs and sources of infections. Most studies on risk factors have focused on multidrug-resistant (MDR) isolates and clinically-oriented questions. Over a two-year period, we sampled 131 wild animals including mammal and bird species from three regions of Central Italy. All typical colonies isolated from the analytical portions were confirmed by real-time PCR and identified by MALDI-TOF mass spectrometry (MALDI-TOF MS). All confirmed K. pneumoniae isolates were tested for antimicrobial susceptibility to 29 antimicrobials and subjected to whole genome sequencing. Typical colonies were detected in 17 samples (13%), which were identified as K. pneumoniae (n = 16) and as K. quasipneumoniae (n = 1) by MALDI-TOF MS. The antimicrobial susceptibility profile showed that all the isolates were resistant to β-lactams (ceftobiprole, cloxacillin, cefazolin) and tetracycline; resistance to ertapenem and trimethoprim was observed and nine out of 16 K. pneumoniae isolates (56.2%) were classified as MDR. Genomic characterization allowed the detection of fluoroquinolone resistance-associated efflux pumps, fosfomycin and β-lactamase resistance genes, and virulence genes in the overall dataset. The cluster analysis of two isolates detected from wild boar with available clinical genomes showed the closest similarity. This study highlights the link between humans, domestic animals, and wildlife, showing that the current knowledge on this ecological context is lacking and that the potential health risks are underestimated.
Keywords: K. pneumoniae; antimicrobial resistance; whole genome sequencing; wildlife.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Radhouani H., Poeta P., Gonçalves A., Pacheco R., Sargo R., Igrejas G. Wild birds as biological indicators of environmental pollution: Antimicrobial resistance patterns of Escherichia coli and enterococci isolated from common buzzards (Buteo buteo) J. Med. Microbiol. 2012;61:837–843. doi: 10.1099/jmm.0.038364-0. - DOI - PubMed
LinkOut - more resources
Full Text Sources

