Genome-Wide DNA Methylation Patterns of Muscle and Tail-Fat in DairyMeade Sheep and Mongolian Sheep
- PMID: 35681863
- PMCID: PMC9179529
- DOI: 10.3390/ani12111399
Genome-Wide DNA Methylation Patterns of Muscle and Tail-Fat in DairyMeade Sheep and Mongolian Sheep
Abstract
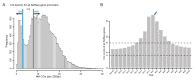
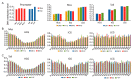
This study aimed to explore the genome-wide DNA methylation differences between muscle and tail-fat tissues of DairyMeade sheep (thin-tailed, lean carcass) and Mongolian sheep (fat-tailed, fat-deposited carcass). Whole-genome bisulfite sequencing (WGBS) was conducted and the global DNA methylation dynamics were mapped. Generally, CGs had a higher DNA methylation level than CHHs and CHGs, and tail-fat tissues had higher CG methylation levels than muscle tissues. For DNA repeat elements, SINE had the highest methylation level, while Simple had the lowest. When dividing the gene promoter region into small bins (200 bp per bin), the bins near the transcription start site (±200 bp) had the highest CG count per bin but the lowest DNA methylation levels. A series of DMRs were identified in muscle and tail-fat tissues between the two breeds. Among them, the introns of gene CAMK2D (calcium/calmodulin-dependent protein kinase II δ) demonstrated significant DNA methylation level differences between the two breeds in both muscle and tail-fat tissues, and it may play a crucial role in fat metabolism and meat quality traits. This study may provide basic datasets and references for further epigenetic modification studies during sheep genetic improvement.
Keywords: CAMK2D; CpG; fat-tailed sheep; gene promoter region; whole-genome bisulfite sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources

