Insights into the Pharmacological Effects of Flavonoids: The Systematic Review of Computer Modeling
- PMID: 35682702
- PMCID: PMC9181432
- DOI: 10.3390/ijms23116023
Insights into the Pharmacological Effects of Flavonoids: The Systematic Review of Computer Modeling
Abstract
Computer modeling is a method that is widely used in scientific investigations to predict the biological activity, toxicity, pharmacokinetics, and synthesis strategy of compounds based on the structure of the molecule. This work is a systematic review of articles performed in accordance with the recommendations of PRISMA and contains information on computer modeling of the interaction of classical flavonoids with different biological targets. The review of used computational approaches is presented. Furthermore, the affinities of flavonoids to different targets that are associated with the infection, cardiovascular, and oncological diseases are discussed. Additionally, the methodology of bias risks in molecular docking research based on principles of evidentiary medicine was suggested and discussed. Based on this data, the most active groups of flavonoids and lead compounds for different targets were determined. It was concluded that flavonoids are a promising object for drug development and further research of pharmacology by in vitro, ex vivo, and in vivo models is required.
Keywords: bias risk; cheminformatics; computer modeling; docking; flavonoids; in silico; limitations; molecular modeling; phytomedicine; systematic review.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

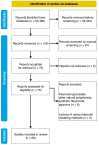
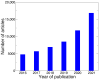


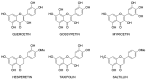

References
-
- Bossel H. Modeling and Simulation. CRC Press; Boca Raton, FL, USA: 2018. pp. 3–6.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

