Evaluation of Potential Anti-Hepatitis A Virus 3C Protease Inhibitors Using Molecular Docking
- PMID: 35682728
- PMCID: PMC9181686
- DOI: 10.3390/ijms23116044
Evaluation of Potential Anti-Hepatitis A Virus 3C Protease Inhibitors Using Molecular Docking
Abstract
Hepatitis A virus (HAV) infection is a major cause of acute hepatitis worldwide and occasionally causes acute liver failure and can lead to death in the absence of liver transplantation. Although HAV vaccination is available, the prevalence of HAV vaccination is not adequate in some countries. Additionally, the improvements in public health reduced our immunity to HAV infection. These situations motivated us to develop potentially new anti-HAV therapeutic options. We carried out the in silico screening of anti-HAV compounds targeting the 3C protease enzyme using the Schrodinger Modeling software from the antiviral library of 25,000 compounds to evaluate anti-HAV 3C protease inhibitors. Additionally, in vitro studies were introduced to examine the inhibitory effects of HAV subgenomic replicon replication and HAV HA11-1299 genotype IIIA replication in hepatoma cell lines using luciferase assays and real-time RT-PCR. In silico studies enabled us to identify five lead candidates with optimal binding interactions in the active site of the target HAV 3C protease using the Schrodinger Glide program. In vitro studies substantiated our hypothesis from in silico findings. One of our lead compounds, Z10325150, showed 47% inhibitory effects on HAV genotype IB subgenomic replicon replication and 36% inhibitory effects on HAV genotype IIIA HA11-1299 replication in human hepatoma cell lines, with no cytotoxic effects at concentrations of 100 μg/mL. The effects of the combination therapy of Z10325150 and RNA-dependent RNA polymerase inhibitor, favipiravir on HAV genotype IB HM175 subgenomic replicon replication and HAV genotype IIIA HA11-1299 replication showed 64% and 48% inhibitory effects of HAV subgenomic replicon and HAV replication, respectively. We identified the HAV 3C protease inhibitor Z10325150 through in silico screening and confirmed the HAV replication inhibitory activity in human hepatocytes. Z10325150 may offer the potential for a useful HAV inhibitor in severe hepatitis A.
Keywords: 3C protease; HAV; in silico screening; molecular docking; protease inhibitors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
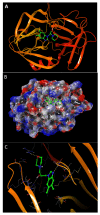



References
-
- Sabrià A., Gregori J., Garcia-Cehic D., Guix S., Pumarola T., Manzanares-Laya S., Caylà J.A., Bosch A., Quer J., Pintó R.M. Evidence for positive selection of hepatitis A virus antigenic variants in vaccinated men-having-sex-with men patients: Implications for immunization policies. EBioMedicine. 2019;39:348–357. doi: 10.1016/j.ebiom.2018.11.023. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

