Endometrial Epithelial ARID1A Is Required for Uterine Immune Homeostasis during Early Pregnancy
- PMID: 35682747
- PMCID: PMC9181301
- DOI: 10.3390/ijms23116067
Endometrial Epithelial ARID1A Is Required for Uterine Immune Homeostasis during Early Pregnancy
Abstract
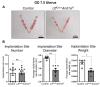
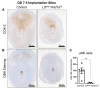
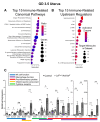
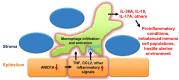
A growing body of work suggests epigenetic dysregulation contributes to endometriosis pathophysiology and female infertility. The chromatin remodeling complex subunit AT-rich interaction domain 1A (ARID1A) must be properly expressed to maintain normal uterine function. Endometrial epithelial ARID1A is indispensable for pregnancy establishment in mice through regulation of endometrial gland function; however, ARID1A expression is decreased in infertile women with endometriosis. We hypothesized that ARID1A performs critical operations in the endometrial epithelium necessary for fertility besides maintaining gland function. To identify alterations in uterine gene expression resulting from loss of epithelial ARID1A, we performed RNA-sequencing analysis on pre-implantation uteri from LtfiCre/+Arid1af/f and control mice. Differential expression analysis identified 4181 differentially expressed genes enriched for immune-related ingenuity canonical pathways including agranulocyte adhesion and diapedesis and natural killer cell signaling. RT-qPCR confirmed an increase in pro-inflammatory cytokine and macrophage-related gene expression but a decrease in natural killer cell signaling. Immunostaining confirmed a uterus-specific increase in macrophage infiltration. Flow cytometry delineated an increase in inflammatory macrophages and a decrease in uterine dendritic cells in LtfiCre/+Arid1af/f uteri. These findings demonstrate a role for endometrial epithelial ARID1A in suppressing inflammation and maintaining uterine immune homeostasis, which are required for successful pregnancy and gynecological health.
Keywords: ARID1A; endometriosis; endometrium; immunology; infertility; inflammation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Endometrial epithelial ARID1A is critical for uterine gland function in early pregnancy establishment.FASEB J. 2021 Feb;35(2):e21209. doi: 10.1096/fj.202002178R. Epub 2020 Nov 22. FASEB J. 2021. PMID: 33222288 Free PMC article.
-
ARID1A Is Essential for Endometrial Function during Early Pregnancy.PLoS Genet. 2015 Sep 17;11(9):e1005537. doi: 10.1371/journal.pgen.1005537. eCollection 2015 Sep. PLoS Genet. 2015. PMID: 26378916 Free PMC article.
-
ARID1A and PGR proteins interact in the endometrium and reveal a positive correlation in endometriosis.Biochem Biophys Res Commun. 2021 Apr 23;550:151-157. doi: 10.1016/j.bbrc.2021.02.144. Epub 2021 Mar 9. Biochem Biophys Res Commun. 2021. PMID: 33706098 Free PMC article.
-
ARID1A mutations and PI3K/AKT pathway alterations in endometriosis and endometriosis-associated ovarian carcinomas.Int J Mol Sci. 2013 Sep 12;14(9):18824-49. doi: 10.3390/ijms140918824. Int J Mol Sci. 2013. PMID: 24036443 Free PMC article. Review.
-
The Role of ARID1A in Endometrial Cancer and the Molecular Pathways Associated With Pathogenesis and Cancer Progression.In Vivo. 2019 May-Jun;33(3):659-667. doi: 10.21873/invivo.11524. In Vivo. 2019. PMID: 31028182 Free PMC article. Review.
Cited by
-
The roles of chromatin regulatory factors in endometriosis.J Assist Reprod Genet. 2024 Apr;41(4):863-873. doi: 10.1007/s10815-024-03026-8. Epub 2024 Jan 25. J Assist Reprod Genet. 2024. PMID: 38270747 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
- R01 HD102170/HD/NICHD NIH HHS/United States
- T32 HD087166/HD/NICHD NIH HHS/United States
- F31HD101207/Eunice Kennedy Shriver National Institute of Child Health and Human Development
- R21 HD099383/HD/NICHD NIH HHS/United States
- R01 HD101243/HD/NICHD NIH HHS/United States
- R01 HD084478/HD/NICHD NIH HHS/United States
- R01HD102170/Eunice Kennedy Shriver National Institute of Child Health and Human Development
- R01HD084478/Eunice Kennedy Shriver National Institute of Child Health and Human Development
- F31 HD101207/HD/NICHD NIH HHS/United States
- T32HD087166/Eunice Kennedy Shriver National Institute of Child Health and Human Development
- R01 HD103617/HD/NICHD NIH HHS/United States
- R01HD101243/Eunice Kennedy Shriver National Institute of Child Health and Human Development
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

