Newly Emerged Antiviral Strategies for SARS-CoV-2: From Deciphering Viral Protein Structural Function to the Development of Vaccines, Antibodies, and Small Molecules
- PMID: 35682761
- PMCID: PMC9181103
- DOI: 10.3390/ijms23116083
Newly Emerged Antiviral Strategies for SARS-CoV-2: From Deciphering Viral Protein Structural Function to the Development of Vaccines, Antibodies, and Small Molecules
Abstract
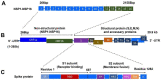
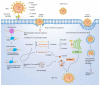
Coronavirus disease 2019 (COVID-19) caused by the infection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become the most severe health crisis, causing extraordinary economic disruption worldwide. SARS-CoV-2 is a single-stranded RNA-enveloped virus. The process of viral replication and particle packaging is finished in host cells. Viral proteins, including both structural and nonstructural proteins, play important roles in the viral life cycle, which also provides the targets of treatment. Therefore, a better understanding of the structural function of virus proteins is crucial to speed up the development of vaccines and therapeutic strategies. Currently, the structure and function of proteins encoded by the SARS-CoV-2 genome are reviewed by several studies. However, most of them are based on the analysis of SARS-CoV-1 particles, lacking a systematic review update for SARS-CoV-2. Here, we specifically focus on the structure and function of proteins encoded by SARS-CoV-2. Viral proteins that contribute to COVID-19 infection and disease pathogenesis are reviewed according to the most recent research findings. The structure-function correlation of viral proteins provides a fundamental rationale for vaccine development and targeted therapy. Then, current antiviral vaccines are updated, such as inactive viral vaccines and protein-based vaccines and DNA, mRNA, and circular RNA vaccines. A summary of other therapeutic options is also reviewed, including monoclonal antibodies such as a cross-neutralizer antibody, a constructed cobinding antibody, a dual functional monoclonal antibody, an antibody cocktail, and an engineered bispecific antibody, as well as peptide-based inhibitors, chemical compounds, and clustered regularly interspaced short palindromic repeats (CRISPR) exploration. Overall, viral proteins and their functions provide the basis for targeted therapy and vaccine development.
Keywords: COVID-19; SARS-CoV-2; antibody treatment; compounds; inhibitors; nonstructural proteins; structural proteins; therapy; vaccines.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Machhi J., Herskovitz J., Senan A.M., Dutta D., Nath B., Oleynikov M.D., Blomberg W.R., Meigs D.D., Hasan M., Patel M., et al. The Natural History, Pathobiology, and Clinical Manifestations of SARS-CoV-2 Infections. J. Neuroimmune Pharmacol. 2020;15:359–386. doi: 10.1007/s11481-020-09944-5. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

