Dissecting the Molecular Regulation of Natural Variation in Growth and Senescence of Two Eutrema salsugineum Ecotypes
- PMID: 35682805
- PMCID: PMC9181637
- DOI: 10.3390/ijms23116124
Dissecting the Molecular Regulation of Natural Variation in Growth and Senescence of Two Eutrema salsugineum Ecotypes
Abstract
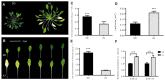
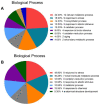
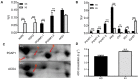
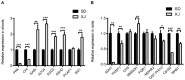
Salt cress (Eutrema salsugineum, aka Thellungiella salsuginea) is an extremophile and a close relative of Arabidopsis thaliana. To understand the mechanism of selection of complex traits under natural variation, we analyzed the physiological and proteomic differences between Shandong (SD) and Xinjiang (XJ) ecotypes. The SD ecotype has dark green leaves, short and flat leaves, and more conspicuous taproots, and the XJ ecotype had greater biomass and showed clear signs of senescence or leaf shedding with age. After 2-DE separation and ESI-MS/MS identification, between 25 and 28 differentially expressed protein spots were identified in shoots and roots, respectively. The proteins identified in shoots are mainly involved in cellular metabolic processes, stress responses, responses to abiotic stimuli, and aging responses, while those identified in roots are mainly involved in small-molecule metabolic processes, oxidation-reduction processes, and responses to abiotic stimuli. Our data revealed the evolutionary differences at the protein level between these two ecotypes. Namely, in the evolution of salt tolerance, the SD ecotype highly expressed some stress-related proteins to structurally adapt to the high salt environment in the Yellow River Delta, whereas the XJ ecotype utilizes the specialized energy metabolism to support this evolution of the short-lived xerophytes in the Xinjiang region.
Keywords: Eutrema salsugineum; growth; molecular regulation; natural variation; salt tolerance; senescence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Inan G., Zhang Q., Li P., Wang Z., Cao Z., Zhang H., Zhang C., Quist T.M., Goodwin S.M., Zhu J., et al. Salt cress. A halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol. 2004;135:1718–1737. doi: 10.1104/pp.104.041723. - DOI - PMC - PubMed
-
- Taji T., Seki M., Satou M., Sakurai T., Kobayashi M., Ishiyama K., Narusaka Y., Narusaka M., Zhu J.K., Shinozaki K. Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol. 2004;135:1697–1709. doi: 10.1104/pp.104.039909. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

