Time-Course Transcriptomic Profiling of Floral Induction in Cultivated Strawberry
- PMID: 35682808
- PMCID: PMC9181015
- DOI: 10.3390/ijms23116126
Time-Course Transcriptomic Profiling of Floral Induction in Cultivated Strawberry
Abstract
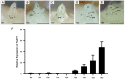
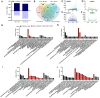
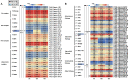
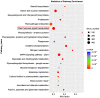
The initiation and quality of flowering directly affect the time to market and economic benefit of cultivated strawberries, but the underlying mechanisms of these processes are largely unknown. To investigate the gene activity during the key period of floral induction in strawberries, time-course transcriptome analysis was performed on the shoot apex of the strawberry cultivar 'Benihoppe.' A total of 7177 differentially expressed genes (DEGs) were identified through pairwise comparisons. These DEGs were grouped into four clusters with dynamic expression patterns. By analyzing the key genes in the potential flowering pathways and the development of the leaf and flower, at least 73 DEGs that may be involved in the regulatory network of floral induction in strawberries were identified, some of which belong to the NAC, MYB, MADS, and SEB families. A variety of eight hormone signaling pathway genes that might play important roles in floral induction were analyzed. In particular, the gene encoding DELLA, a key inhibitor of the gibberellin signaling pathway, was found to be significantly differentially expressed during the floral induction. Furthermore, the differential expression of some important candidate genes, such as TFL1, SOC1, and GAI-like, was further verified by qRT-PCR. Therefore, we used this time-course transcriptome data for a preliminary exploration of the regulatory network of floral induction and to provide potential candidate genes for future studies of flowering in strawberries.
Keywords: RNA-seq; floral induction; hormone; strawberry; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Hytönen T., Elomaa P. Genetic and Environmental Regulation of Flowering and Runnering in Strawberry. Genes Genomes Genom. 2011;5:56–64.
-
- Jonkers H. Ph.D. Thesis. Mededelingen van de Landbouwhogeschool; Wageningen, The Netherlands: 1965. On the Flower Formation, the Dormancy and the Early Forcing of Strawberries.
-
- Heide O.M. Photoperiod and Temperature Interactions in Growth and Flowering of Strawberry. Physiol. Plantarum. 1977;40:21–26. doi: 10.1111/j.1399-3054.1977.tb01486.x. - DOI
-
- Heide O.M., Sønsteby A. Interactions of temperature and photoperiod in the control of flowering of latitudinal and altitudinal populations of wild strawberry (Fragaria vesca) Physiol. Plant. 2010;130:280–289. doi: 10.1111/j.1399-3054.2007.00906.x. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

