Intestinal Mucosal Barrier Improvement with Prebiotics: Histological Evaluation of Longish Glucomannan Hydrolysates-Induced Innate T Lymphocyte Activities in Mice
- PMID: 35684019
- PMCID: PMC9182621
- DOI: 10.3390/nu14112220
Intestinal Mucosal Barrier Improvement with Prebiotics: Histological Evaluation of Longish Glucomannan Hydrolysates-Induced Innate T Lymphocyte Activities in Mice
Abstract
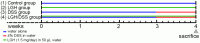
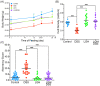
Use of prebiotics is a growing topic in healthcare. A lightweight molecule and water-soluble fiber ingredient, longish glucomannan hydrolysates (LGH), has been developed to improve the intestinal mucosal barrier and confer gut health benefits. This study aims to investigate the implications of continuous LGH intervening in intestinal epithelium integrity and protective immunity against chemical dextran sodium sulfate (DSS)-induced colitis. Twelve male BALB/c mice were randomly arranged into four groups. The LGH/DSS group had results in bodyweight variance, epithelial cell density, and aberrancy score as good as the LGH group, and both were equivalent to the control group. LGH consumption effectively protects the distal intestinal epithelium by activating innate T lymphocytes. Meanwhile, T-cell subsets in subepithelial interspersion take a bystander role in these microenvironmental alterations. Under this stress, the cluster of differentiation 3 (CD3)+ T cells infiltrate the epithelium, while CD4+ T cells inversely appear in submucosal large lymphoid aggregates/isolated lymphoid follicles (ILFs) in which significant CD3+, CD4+, and CD8+ T-cell populations agglomerate. Moreover, forkhead box P3 (Foxp3) and interleukin 17 (IL-17) are observed in these ILFs. Agglomerated CD4+ T-cell lineages may have roles with proinflammatory T helper 17 cells and anti-inflammatory regulatory T cells in balancing responses to intraluminal antigens. Collectively, LGH administration may function in immune modulation to protect against DSS-induced inflammation.
Keywords: T-cell activation; colitis; glucomannan; lymphoid aggregates; prebiotics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

