Transcriptome Profiling of HCT-116 Colorectal Cancer Cells with RNA Sequencing Reveals Novel Targets for Polyphenol Nano Curcumin
- PMID: 35684411
- PMCID: PMC9182402
- DOI: 10.3390/molecules27113470
Transcriptome Profiling of HCT-116 Colorectal Cancer Cells with RNA Sequencing Reveals Novel Targets for Polyphenol Nano Curcumin
Abstract
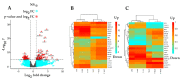
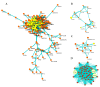
Colorectal cancer is one of the leading causes of cancer-related deaths worldwide. The gemini nanoparticle formulation of polyphenolic curcumin significantly inhibits the viability of cancer cells. However, the molecular mechanisms and pathways underlying its toxicity in colon cancer are unclear. Here, we aimed to uncover the possible novel targets of gemini curcumin (Gemini-Cur) on colorectal cancer and related cellular pathways. After confirming the cytotoxic effect of Gemini-Cur by MTT and apoptotic assays, RNA sequencing was employed to identify differentially expressed genes (DEGs) in HCT-116 cells. On a total of 3892 DEGs (padj < 0.01), 442 genes showed a log2 FC >|2| (including 244 upregulated and 198 downregulated). Gene ontology (GO) enrichment analysis was performed. Protein−protein interaction (PPI) and gene-pathway networks were constructed by using STRING and Cytoscape. The pathway analysis showed that Gemini-Cur predominantly modulates pathways related to the cell cycle. The gene network analysis revealed five central genes, namely GADD45G, ATF3, BUB1B, CCNA2 and CDK1. Real-time PCR and Western blotting analysis confirmed the significant modulation of these genes in Gemini-Cur-treated compared to non-treated cells. In conclusion, RNA sequencing revealed novel potential targets of curcumin on cancer cells. Further studies are required to elucidate the molecular mechanism of action of Gemini-Cur regarding the modulation of the expression of hub genes.
Keywords: PPI network; RNA sequencing; colorectal cancer; differentially expressed genes; gemini curcumin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Guo Y., Wu R., Gaspar J.M., Sargsyan D., Su Z.-Y., Zhang C., Gao L., Cheng D., Li W., Wang C., et al. DNA methylome and transcriptome alterations and cancer prevention by curcumin in colitis-accelerated colon cancer in mice. Carcinogenesis. 2018;39:669–680. doi: 10.1093/carcin/bgy043. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

