Fine Mapping of Rice Specific MR1, a Gene Determines Palea Identity
- PMID: 35685009
- PMCID: PMC9171376
- DOI: 10.3389/fpls.2022.864099
Fine Mapping of Rice Specific MR1, a Gene Determines Palea Identity
Abstract
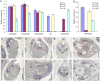
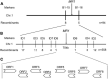
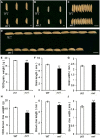
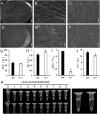
The hull (palea and lemma) is the specific organ of grass florets. Although many genes related to the hull development have been cloned, the genetic mechanisms behind the development are still unclear, and the evolutionary relationship has different explanations and heated arguments between the palea and lemma. In this study, we found a specific mr1 mutant with a reduced palea, showing an enlarged mrp and degraded bop. Phenotype observations and molecular evidences showed that the bop was converted to the mrp-like organ. Our findings first reveal that the bop and mrp are homologous structures, and the palea and lemma are the same whorl floral organs. MR1 may prevent the transformation of the bop into mrp by regulating the expressions of hull identity genes. Meantime, the mr1 mutant showed altered grain size and grain quality, with defective physical and chemical contents. MR1 was controlled by a single recessive gene and was finally located on chromosome 1, with a physical distance of 70 kb. More work will be needed for confirming the target gene of MR1, which would contribute to our understanding of grain formation and the origin between the lemma, bop, and mrp.
Keywords: grain size and quality; mr1 mutant; organ origin; palea; rice (Oryza sativa L.).
Copyright © 2022 Xie, Liu, Yu, Zeng and Ren.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources

