Integrative RNA profiling of TBEV-infected neurons and astrocytes reveals potential pathogenic effectors
- PMID: 35685361
- PMCID: PMC9167876
- DOI: 10.1016/j.csbj.2022.05.052
Integrative RNA profiling of TBEV-infected neurons and astrocytes reveals potential pathogenic effectors
Abstract
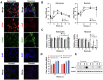
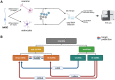
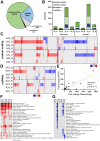
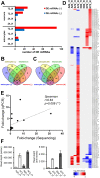
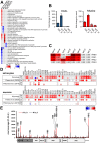
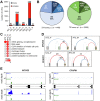
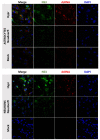
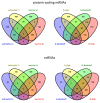
Tick-borne encephalitis virus (TBEV), the most medically relevant tick-transmitted flavivirus in Eurasia, targets the host central nervous system and frequently causes severe encephalitis. The severity of TBEV-induced neuropathogenesis is highly cell-type specific and the exact mechanism responsible for such differences has not been fully described yet. Thus, we performed a comprehensive analysis of alterations in host poly-(A)/miRNA/lncRNA expression upon TBEV infection in vitro in human primary neurons (high cytopathic effect) and astrocytes (low cytopathic effect). Infection with severe but not mild TBEV strain resulted in a high neuronal death rate. In comparison, infection with either of TBEV strains in human astrocytes did not. Differential expression and splicing analyses with an in silico prediction of miRNA/mRNA/lncRNA/vd-sRNA networks found significant changes in inflammatory and immune response pathways, nervous system development and regulation of mitosis in TBEV Hypr-infected neurons. Candidate mechanisms responsible for the aforementioned phenomena include specific regulation of host mRNA levels via differentially expressed miRNAs/lncRNAs or vd-sRNAs mimicking endogenous miRNAs and virus-driven modulation of host pre-mRNA splicing. We suggest that these factors are responsible for the observed differences in the virulence manifestation of both TBEV strains in different cell lines. This work brings the first complex overview of alterations in the transcriptome of human astrocytes and neurons during the infection by two TBEV strains of different virulence. The resulting data could serve as a starting point for further studies dealing with the mechanism of TBEV-host interactions and the related processes of TBEV pathogenesis.
Keywords: A3SS, alternative 3′ splice site; A5SS, alternative 5′ splice site; ACACA, Acetyl-CoA Carboxylase Alpha; AKR1C2, Aldo-Keto Reductase Family 1 Member C2; ANKS1A, Ankyrin Repeat And Sterile Alpha Motif Domain Containing 1A; ANOS1, Anosmin 1; AOX1, Aldehyde Oxidase 1; APOBEC3G, Apolipoprotein B MRNA Editing Enzyme Catalytic Subunit 3G; APOL1/6, Apolipoprotein L1/6; ARID2, AT-Rich Interaction Domain 2; AUTS2, Activator Of Transcription And Developmental Regulator AUTS2; Alternative splicing; Astrocytes; BCL11B, BAF Chromatin Remodeling Complex Subunit BCL11B; BCL9L, BCL9 Transcription Coactivator-like; BDKRB2, Bradykinin Receptor B2; BDNF, Brain Derived Neurotrophic Factor; BEND3, BEN Domain Containing 3; BSA, bovine serum albumin; BST2, Bone Marrow Stromal Cell Antigen 2; CALB1, Calbindin 1; CAMK2A, Calcium/Calmodulin Dependent Protein Kinase II Alpha; CD, complement determinant; CDKN1C, Cyclin Dependent Kinase Inhibitor 1C; CFAP61, Cilia And Flagella Associated Protein 61; CHRNA3, Cholinergic Receptor Nicotinic Alpha 3 Subunit; CHRNB4, Cholinergic Receptor Nicotinic Beta 4 Subunit; CLIC5, Chloride Intracellular Channel 5; CMPK2, Cytidine/Uridine Monophosphate Kinase 2; CNS, central nervous system; CNTN2, Contactin 2; CREG2, Cellular Repressor Of E1A Stimulated Genes 2; CXADR, Coxsackievirus B-Adenovirus Receptor; CYYR1, Cysteine And Tyrosine Rich 1; DACH1, Dachshund Family Transcription Factor 1; DAPI, diamidino-2-phenylindole; DCC, Netrin 1 Receptor; DCX, Doublecortin; DDX60, DExD/H-Box Helicase 60; DDX60L, DExD/H-Box 60 Like; DE, differentially expressed; DENV, Dengue virus; DIRAS2, DIRAS Family GTPase 2; DLX1/5/6, Distal-Less Homeobox 1/5/6; DNMT3B, DNA Methyltransferase 3 Beta; DPYSL2, Dihydropyrimidinase Like 2; EBF1, EBF Transcription Factor 1; EGF, Epidermal Growth Factor; ELAVL2/4, ELAV Like RNA Binding Protein 2/4; EPHB1, EPH Receptor B1; EPSTI1, Epithelial Stromal Interaction 1; ERBB4, Erb-B2 Receptor Tyrosine Kinase 4; ES, exon skipping; ESRRG, Estrogen Related Receptor Gamma; FGFb, Fibroblast Growth Factor 2; FPKM, Fragments Per Kilobase of transcript per Million mapped reads; FUT9, Fucosyltransferase 9; G2E3, G2/M−Phase Specific E3 Ubiquitin Protein Ligase; GABRG2, Gamma-Aminobutyric Acid Type A Receptor Subunit Gamma 2; GAPDH, Glyceraldehyde-3-Phosphate Dehydrogenase; GAS2L3, Growth Arrest Specific 2 Like 3; GAS7, Growth Arrest Specific 7; GATAD2B, GATA Zinc Finger Domain Containing 2B; GFAP, Glial Fibrillary Acidic Protein; GIPC2, GIPC PDZ Domain Containing Family Member 2; GLRA2, Glycine Receptor Alpha 2; GNG2, G Protein Subunit Gamma 2; GO, gene ontology; GOLGA4, Golgin A4; GRIN2A, Glutamate Ionotropic Receptor NMDA Type Subunit 2A; GSEA, gene set enrichment analysis; HERC5/6, HECT And RLD Domain Containing E3 Ubiquitin Protein Ligase 5/6; HEYL, Hes Related Family BHLH Transcription Factor With YRPW Motif Like; HPRT1, Hypoxanthine Phosphoribosyltransferase 1; HS, hot-spot; HSPA6, Heat Shock Protein Family A (Hsp70) Member 6; HUDD (ELAV4), Hu-Antigen D/ELAV Like Neuron-Specific RNA Binding Protein 4; IFI6, Interferon Alpha Inducible Protein 6; IFIH1 (MDA5), Interferon Induced With Helicase C Domain 1/Melanoma Differentiation-Associated Protein 5; IFIT1-3, Interferon Induced Protein With Tetratricopeptide Repeats 1–3; IFITM1/2, Interferon Induced Transmembrane Protein 1/2; IFN, interferon; IGB, Integrated Genome Browser; IL6, Interleukin 6; IR, intron retention; ISG20, Interferon Stimulated Exonuclease Gene 20; ISGF3, Interferon-Stimulated Gene Factor 3 Gamma; ISGs, interferon-stimulated genes; JEV, Japanese encephalitis virus; KCND2, Potassium Voltage-Gated Channel Subfamily D Member 2; KCNK10, Potassium Two Pore Domain Channel Subfamily K Member 10; KCNS2, Potassium Voltage-Gated Channel Modifier Subfamily S Member 2; KIT, KIT Proto-Oncogene, Receptor Tyrosine Kinase; KLHDC8A, Kelch Domain Containing 8A; KLHL13, Kelch Like Family Member 13; KRR1, KRR1 Small Subunit Processome Component Homolog; LCOR, Ligand Dependent Nuclear Receptor Corepressor; LEKR1, Leucine, Glutamate And Lysine Rich 1; LGI1, Leucine Rich Glioma Inactivated 1; LRRTM3, Leucine Rich Repeat Transmembrane Neuronal 3; LSV, local splicing variation; LUZP2, Leucine Zipper Protein 2; MAN1A1, Mannosidase Alpha Class 1A Member 1; MAP2, Microtubule Associated Protein 2; MBNL2, Muscleblind Like Splicing Regulator 2; MCTP1, Multiple C2 And Transmembrane Domain Containing 1; MMP13, Matrix Metallopeptidase 13; MN1, MN1 Proto-Oncogene, Transcriptional Regulator; MOI, multiplicity of infection; MTUS2, Microtubule Associated Scaffold Protein 2; MX2, MX Dynamin Like GTPase 2; MYCN, MYCN Proto-Oncogene, BHLH Transcription Factor; NAV1, Neuron Navigator 1; NCAM1, Neural Cell Adhesion Molecule 1; NDRG4, N-Myc Downstream-Regulated Gene 4 Protein; NEK7, NIMA Related Kinase 7; NFASC, Neurofascin; NKAIN1, Sodium/Potassium Transporting ATPase Interacting 1; NMI, N-Myc And STAT Interactor 2; NRAP, Nebulin Related Anchoring Protein; NRARP, NOTCH Regulated Ankyrin Repeat Protein; NREP, Neuronal Regeneration Related Protein; NRN1, Neuritin 1; NS3, flaviviral non-structural protein 3; NXPH2, Neurexophilin 2; NYNRIN, NYN Domain And Retroviral Integrase Containing; Neurons; Neuropathogenesis; OAS, 2′-5′-Oligoadenylate Synthetase; OASL, 2′-5′-Oligoadenylate Synthetase Like; ONECUT2, ONECUT-2 Homeodomain Transcription Factor; OPCML, Opioid Binding Protein/Cell Adhesion Molecule Like; OTX2, Orthodenticle Homeobox 2; PBS, phosphate buffer saline; PBX1, Pre-B-Cell Leukemia Transcription Factor 1; PCDH18/20, Protocadherin 18/20; PFKFB3, 6-Phosphofructo-2-Kinase/Fructose-2,6-Biphosphatase 3; PIK3C2B, Phosphatidylinositol-4-Phosphate 3-Kinase Catalytic Subunit Type 2 Beta; PIP4P2, Phosphatidylinositol-4,5-Bisphosphate 4-Phosphatase 2; PLCH1, Phospholipase C Eta 1; POU3F4, Brain-Specific Homeobox/POU Domain Protein 4; PPM1L, Protein Phosphatase, Mg2+/Mn2+ Dependent 1L; PPP1R17, Protein Phosphatase 1 Regulatory Subunit 17; PRDM12, PR Domain Zinc Finger Protein 12; PSI, percent selective index; PSRC1, Proline And Serine Rich Coiled-Coil 1; PTPN5, Protein Tyrosine Phosphatase Non-Receptor Type 5; PTPRH, Protein Tyrosine Phosphatase Receptor Type H; RAPGEF5, Rap Guanine Nucleotide Exchange Factor 5; RBFOX1, RNA Binding Fox-1 Homolog 1; RIG-I (DDX58), Retinoic Acid-Inducible Gene 1 Protein; RNF212, Ring Finger Protein 212; RNVU1, RNA, Variant U1 Small Nuclear; RSAD2, Radical S-Adenosyl Methionine Domain Containing 2; RTL8B, Retrotransposon Gag Like 8B; Response to infection; SAMD9, Sterile Alpha Motif Domain Containing 9; SEMA3E, Semaphorin 3E; SH3TC2, SH3 Domain And Tetratricopeptide Repeats 2; SHF, Src Homology 2 Domain Containing F; SHISAL1, Shisa Like 1; SIAH3, Siah E3 Ubiquitin Protein Ligase Family Member 3; SIRPA, Signal Regulatory Protein Alpha; SLITRK5, SLIT And NTRK Like Family Member 5; SNP, single-nucleotide polymorphism; SOGA1, Suppressor Of Glucose, Autophagy Associated 1; SPSB4, SplA/Ryanodine Receptor Domain And SOCS Box Containing 4; ST6GAL1, ST6 Beta-Galactoside Alpha-2,6-Sialyltransferase 1; TBC1D30, TBC1 Domain Family Member 30; TBEV, Tick-borne encephalitis virus; TFAP2A, Transcription Factor AP-2 Alpha; TFAP2B, Transcription Factor AP-2 Beta; THSD7A, Thrombospondin Type 1 Domain Containing 7A; THUMPD2, THUMP Domain-Containing Protein 2/SAM-Dependent Methyltransferase; TIPARP, TCDD Inducible Poly(ADP-Ribose) Polymerase; TM4SF18, Transmembrane 4 L Six Family Member 18; TMC8, Transmembrane Channel Like 6; TMEM229B, Transmembrane Protein 229B; TMTC1, Transmembrane O-Mannosyltransferase Targeting Cadherins 1; TNFSF10, TNF Superfamily Member 10; TRHDE, Thyrotropin Releasing Hormone Degrading Enzyme; TRIM38, Tripartite Motif Containing 38; TSHZ1, Teashirt Zinc Finger Homeobox 1; Tick-borne encephalitis virus; Transcriptomics; USP18, Ubiquitin Specific Peptidase 18/ISG15-Specific-Processing Protease; UTR, untranslated region; UTS2R, Urotensin 2 Receptor; WNV, West Nile virus; XAF1, XIAP Associated Factor 1; XRN1, 5′-3′ Exoribonuclease 1; ZIKV, Zika virus; ZMAT3, Zinc Finger Matrin-Type 3; ZMYM5, Zinc Finger MYM-Type Containing 5; ZNF124, Zinc Finger Protein 124; ZNF730, Zinc Finger Protein 730; gRNA, genomic TBEV RNA; hNSC, human neural stem cells; lncRNA, long non-coding RNA; mRNA, messenger RNA; miRNA; miRNA, micro RNA; ncRNA, non-coding RNA; pc-mRNA, protein-coding mRNA; qRT-PCR, quantitative reverse transcription real-time PCR; snRNP, small nuclear ribonucleoproteins; vd-sRNA, virus-derived small RNA.
© 2022 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


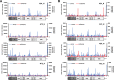
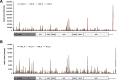

References
-
- Gritsun TS, Lashkevich VA, Gould EA. Tick-borne encephalitis. Antiviral research. 2003;57(1-2):129-46. Epub 2003/03/05. doi: 10.1016/s0166-3542(02)00206-PubMed PMID: 12615309. - PubMed
-
- de Graaf JA, Reimerink JH, Voorn GP, Bij de Vaate EA, de Vries A, Rockx B, et al. First human case of tick-borne encephalitis virus infection acquired in the Netherlands, July 2016. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2016;21(33). Epub 2016/08/27. doi: 10.2807/1560-7917.Es .2016.21.33.30318. PubMed PMID: 27562931; PubMed Central PMCID: PMCPMC4998423. - PMC - PubMed
-
- Gelpi E, Preusser M, Garzuly F, Holzmann H, Heinz FX, Budka H. Visualization of Central European tick-borne encephalitis infection in fatal human cases. Journal of neuropathology and experimental neurology. 2005;64(6):506-12. Epub 2005/06/28. doi: 10.1093/jnen/64.6.506. PubMed PMID: 15977642. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
