Non-steroidal anti-inflammatory drugs: recent advances in the use of synthetic COX-2 inhibitors
- PMID: 35685617
- PMCID: PMC9132194
- DOI: 10.1039/d1md00280e
Non-steroidal anti-inflammatory drugs: recent advances in the use of synthetic COX-2 inhibitors
Abstract
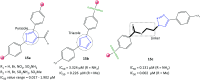
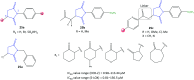
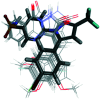
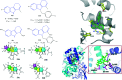
Cyclooxygenase (COX) enzymes comprise COX-1 and COX-2 isoforms and are responsible for prostaglandin production. Prostaglandins have critical roles in the inflammation pathway and must be controlled by administration of selective nonsteroidal anti-inflammatory drugs (NSAIDs). Selective COX-2 inhibitors have been among the most used NSAIDs during the ongoing coronavirus 2019 pandemic because they reduce pain and protect against inflammation-related diseases. In this framework, the mechanism of action of both COX isoforms (particularly COX-2) as inflammation mediators must be reviewed. Moreover, proinflammatory cytokines such as tumor necrosis factor-α and interleukin (IL)-6, IL-1β, and IL-8 must be highlighted due to their major participation in upregulation of the inflammatory reaction. Structural and functional analyses of selective COX-2 inhibitors within the active-site cavity of COXs could enable introduction of lead structures with higher selectivity and potency against inflammation with fewer adverse effects. This review focuses on the biological activity of recently discovered synthetic COX-2, dual COX-2/lipoxygenase, and COX-2/soluble epoxide hydrolase hybrid inhibitors based primarily on the active motifs of related US Food and Drug Administration-approved drugs. These new agents could provide several advantages with regard to anti-inflammatory activity, gastrointestinal protection, and a safer profile compared with those of the NSAIDs celecoxib, valdecoxib, and rofecoxib.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




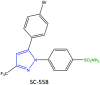

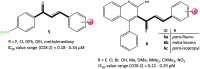


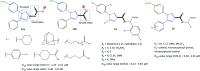
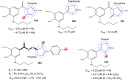










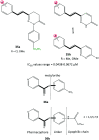
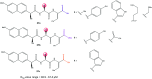










References
Publication types
LinkOut - more resources
Full Text Sources
Chemical Information
Research Materials

