Carboxypeptidase N2 as a Novel Diagnostic and Prognostic Biomarker for Lung Adenocarcinoma
- PMID: 35686102
- PMCID: PMC9170673
- DOI: 10.3389/fonc.2022.843325
Carboxypeptidase N2 as a Novel Diagnostic and Prognostic Biomarker for Lung Adenocarcinoma
Abstract
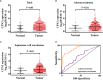
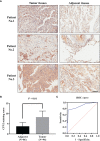
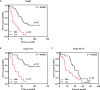
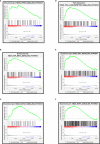
Carboxypeptidase N2 (CPN2) is a plasma metallo-protease that cleaves basic amino acids from the C-terminal of peptides and proteins. Emerging evidence showed that carboxypeptidases perform many diverse functions in the body and play key roles in tumorigenesis. However, the clinical significance and biological functions of CPN2 in lung adenocarcinoma remain unclear. Our study aimed to explore the potential role and functions of CPN2 in lung adenocarcinoma. The results showed that the transcription level of CPN2 was significantly increased in the tumor tissues of lung adenocarcinoma patients compared to the adjacent normal tissues in The Cancer Genome Atlas cohort (P < 0.05). The survival plots showed that the overall survival of patients with a high expression of CPN2 was significantly lower than that of patients with a low expression of CPN2, both in the Kaplan-Meier database and the clinical sample cohort (P < 0.05). The tissue microarray analysis found that CPN2 protein expression was significantly positively correlated with node status and tumor stage as well as tumor malignancy (P < 0.05). Further univariate and multivariate Cox regression analyses showed that CPN2 may act as an independent prognostic factor in patients with lung adenocarcinoma (P < 0.05). In addition, the analysis of co-expression genes from LinkedOmics showed that CPN2 was positively associated with many genes of fibrillar collagen family members and the PI3K-Akt pathway. The gene set enrichment analysis showed that a higher expression of CPN2 may participate in mTOR, TGF-BETA, NOTCH, TOLL-like-receptor, WNT, and MAPK signaling pathway in lung adenocarcinoma. Notably, the knockdown of CPN2 significantly inhibited the ability of cell proliferation, clone formation, invasion, and migration. Our findings suggested that the upregulation of CPN2 is associated with a worse clinical outcome in lung adenocarcinoma and cancer-related pathways, which laid the foundation for further research on CPN2 during carcinogenesis.
Keywords: CPN2; biomarker; diagnosis; lung adenocarcinoma; prognosis.
Copyright © 2022 Xu, Zhang, Chen, Cai, Yang, Liu, Fan, Liu and Yao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
TEAD4 functions as a prognostic biomarker and triggers EMT via PI3K/AKT pathway in bladder cancer.J Exp Clin Cancer Res. 2022 May 17;41(1):175. doi: 10.1186/s13046-022-02377-3. J Exp Clin Cancer Res. 2022. PMID: 35581606 Free PMC article.
-
High expression of RRM2 as an independent predictive factor of poor prognosis in patients with lung adenocarcinoma.Aging (Albany NY). 2020 Dec 19;13(3):3518-3535. doi: 10.18632/aging.202292. Epub 2020 Dec 19. Aging (Albany NY). 2020. PMID: 33411689 Free PMC article.
-
Expression profile, clinical significance, and biological function of insulin-like growth factor 2 messenger RNA-binding proteins in non-small cell lung cancer.Tumour Biol. 2017 Apr;39(4):1010428317695928. doi: 10.1177/1010428317695928. Tumour Biol. 2017. PMID: 28381175
-
Overexpression of hsa_circ_0001715 is a Potential Diagnostic and Prognostic Biomarker in Lung Adenocarcinoma.Onco Targets Ther. 2020 Oct 23;13:10775-10783. doi: 10.2147/OTT.S274932. eCollection 2020. Onco Targets Ther. 2020. PMID: 33122916 Free PMC article.
-
Upregulation of desmoglein 2 and its clinical value in lung adenocarcinoma: a comprehensive analysis by multiple bioinformatics methods.PeerJ. 2020 Feb 13;8:e8420. doi: 10.7717/peerj.8420. eCollection 2020. PeerJ. 2020. PMID: 32095325 Free PMC article.
Cited by
-
Genetic underpinnings explored: OPA1 deletion and complex phenotypes on chromosome 3q29.BMC Med Genomics. 2024 Apr 19;17(1):94. doi: 10.1186/s12920-024-01850-6. BMC Med Genomics. 2024. PMID: 38641846 Free PMC article.
-
Properdin, transcortin and HGFAC are novel plasma biomarkers in canine chronic inflammatory enteropathies from active disease to remission.Sci Rep. 2025 Jul 15;15(1):25443. doi: 10.1038/s41598-025-11474-0. Sci Rep. 2025. PMID: 40659816 Free PMC article.
-
Discovering Breast Cancer Biomarkers Candidates through mRNA Expression Analysis Based on The Cancer Genome Atlas Database.J Pers Med. 2022 Oct 21;12(10):1753. doi: 10.3390/jpm12101753. J Pers Med. 2022. PMID: 36294892 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

