Early Detection of Severe Acute Respiratory Syndrome Coronavirus 2 Variants Using Traveler-based Genomic Surveillance at 4 US Airports, September 2021-January 2022
- PMID: 35686436
- PMCID: PMC9214179
- DOI: 10.1093/cid/ciac461
Early Detection of Severe Acute Respiratory Syndrome Coronavirus 2 Variants Using Traveler-based Genomic Surveillance at 4 US Airports, September 2021-January 2022
Abstract
We enrolled arriving international air travelers in a severe acute respiratory syndrome coronavirus 2 genomic surveillance program. We used molecular testing of pooled nasal swabs and sequenced positive samples for sublineage. Traveler-based surveillance provided early-warning variant detection, reporting the first US Omicron BA.2 and BA.3 in North America.
Keywords: SARS-CoV-2; genomic surveillance; international travelers.
© The Author(s) 2022. Published by Oxford University Press on behalf of Infectious Diseases Society of America.
Conflict of interest statement
Potential conflicts of interest. R. D. W., R. M., S. L. L., K. R., D. D., B. P. G., A. L. H., B. C., G. W., A. M. P., and B. B. S. are employed by Ginkgo Bioworks and own Ginkgo Bioworks employee stocks and/or Restricted Stock Units (RSU) grants. S. R. M., E. T. E., and W. W. D. are employed by the XpresSpa Group and own employee stocks and/or RSU grants. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Figures
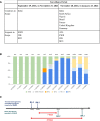
References
-
- TaqPath COVID-19 Tests Support Detection of SARS-CoV-2 in Samples Containing the Omicron Variant. Available at: https://www.thermofisher.com/blog/clinical-conversations/taqpath-covid-1.... Accessed 21 December 2021.
-
- Tyson JR, James P, Stoddart D, et al. . Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. bioRxiv 2020.
-
- Centers for Disease Control and Prevention . How to report COVID-19 laboratory data. Available at: https://www.cdc.gov/coronavirus/2019-ncov/lab/reporting-lab-data.html. Accessed 28 December 2021.
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

