Sequence and structural conservation reveal fingerprint residues in TRP channels
- PMID: 35686986
- PMCID: PMC9242649
- DOI: 10.7554/eLife.73645
Sequence and structural conservation reveal fingerprint residues in TRP channels
Abstract
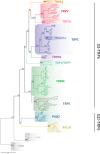
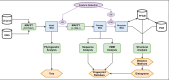
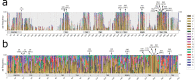
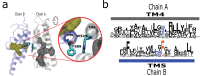
Transient receptor potential (TRP) proteins are a large family of cation-selective channels, surpassed in variety only by voltage-gated potassium channels. Detailed molecular mechanisms governing how membrane voltage, ligand binding, or temperature can induce conformational changes promoting the open state in TRP channels are still a matter of debate. Aiming to unveil distinctive structural features common to the transmembrane domains within the TRP family, we performed phylogenetic reconstruction, sequence statistics, and structural analysis over a large set of TRP channel genes. Here, we report an exceptionally conserved set of residues. This fingerprint is composed of twelve residues localized at equivalent three-dimensional positions in TRP channels from the different subtypes. Moreover, these amino acids are arranged in three groups, connected by a set of aromatics located at the core of the transmembrane structure. We hypothesize that differences in the connectivity between these different groups of residues harbor the apparent differences in coupling strategies used by TRP subgroups.
Keywords: MSA; TRP channels; allosterism; evolution; evolutionary biology; structure.
© 2022, Cabezas-Bratesco, Mcgee et al.
Conflict of interest statement
DC, FM, CC, KZ, DG, VC, JO, SB No competing interests declared
Figures














Similar articles
-
Structural and Evolutionary Insights Point to Allosteric Regulation of TRP Ion Channels.Acc Chem Res. 2019 Jun 18;52(6):1643-1652. doi: 10.1021/acs.accounts.9b00075. Epub 2019 May 31. Acc Chem Res. 2019. PMID: 31149807 Free PMC article.
-
Yeast gain-of-function mutations reveal structure-function relationships conserved among different subfamilies of transient receptor potential channels.Proc Natl Acad Sci U S A. 2007 Dec 4;104(49):19607-12. doi: 10.1073/pnas.0708584104. Epub 2007 Nov 27. Proc Natl Acad Sci U S A. 2007. PMID: 18042709 Free PMC article.
-
The transient receptor potential family of ion channels.Genome Biol. 2011;12(3):218. doi: 10.1186/gb-2011-12-3-218. Epub 2011 Mar 17. Genome Biol. 2011. PMID: 21401968 Free PMC article. Review.
-
Comparative sequence analysis suggests a conserved gating mechanism for TRP channels.J Gen Physiol. 2015 Jul;146(1):37-50. doi: 10.1085/jgp.201411329. Epub 2015 Jun 15. J Gen Physiol. 2015. PMID: 26078053 Free PMC article.
-
Structural mechanisms of transient receptor potential ion channels.J Gen Physiol. 2020 Mar 2;152(3):e201811998. doi: 10.1085/jgp.201811998. J Gen Physiol. 2020. PMID: 31972006 Free PMC article. Review.
Cited by
-
Progress in the Structural Basis of thermoTRP Channel Polymodal Gating.Int J Mol Sci. 2023 Jan 1;24(1):743. doi: 10.3390/ijms24010743. Int J Mol Sci. 2023. PMID: 36614186 Free PMC article. Review.
-
Mechanoresponses mediated by the TRP11 channel in cilia of Chlamydomonas reinhardtii.iScience. 2023 Sep 15;26(10):107926. doi: 10.1016/j.isci.2023.107926. eCollection 2023 Oct 20. iScience. 2023. PMID: 37790279 Free PMC article.
-
The chemistry of electrical signaling in sodium channels from bacteria and beyond.Cell Chem Biol. 2024 Aug 15;31(8):1405-1421. doi: 10.1016/j.chembiol.2024.07.010. Cell Chem Biol. 2024. PMID: 39151407 Free PMC article. Review.
-
Conservation of the cooling agent binding pocket within the TRPM subfamily.Elife. 2024 Nov 1;13:RP99643. doi: 10.7554/eLife.99643. Elife. 2024. PMID: 39485376 Free PMC article.
-
Defining the Polycystin Pharmacophore Through HTS & Computational Biophysics.bioRxiv [Preprint]. 2025 Jan 17:2025.01.13.632808. doi: 10.1101/2025.01.13.632808. bioRxiv. 2025. Update in: Br J Pharmacol. 2025 Oct;182(19):4611-4624. doi: 10.1111/bph.70080. PMID: 39868095 Free PMC article. Updated. Preprint.
References
-
- Altenhoff AM, Glover NM, Train C-M, Kaleb K, Warwick Vesztrocy A, Dylus D, de Farias TM, Zile K, Stevenson C, Long J, Redestig H, Gonnet GH, Dessimoz C. The OMA orthology database in 2018: retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Research. 2018;46:D477–D485. doi: 10.1093/nar/gkx1019. - DOI - PMC - PubMed
-
- Altenhoff AM, Train C-M, Gilbert KJ, Mediratta I, Mendes de Farias T, Moi D, Nevers Y, Radoykova H-S, Rossier V, Warwick Vesztrocy A, Glover NM, Dessimoz C. OMA orthology in 2021: website overhaul, conserved isoforms, ancestral gene order and more. Nucleic Acids Research. 2021;49:D373–D379. doi: 10.1093/nar/gkaa1007. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

