Exploring the Conformational and Binding Dynamics of HMGA2·DNA Complexes Using Trapped Ion Mobility Spectrometry-Mass Spectrometry
- PMID: 35687119
- PMCID: PMC9280850
- DOI: 10.1021/jasms.2c00101
Exploring the Conformational and Binding Dynamics of HMGA2·DNA Complexes Using Trapped Ion Mobility Spectrometry-Mass Spectrometry
Abstract
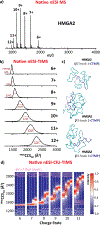
The mammalian high mobility group protein AT-hook 2 (HMGA2) is an intrinsically disordered DNA-binding protein expressed during embryogenesis. In the present work, the conformational and binding dynamics of HMGA2 and HMGA2 in complex with a 22-nt (DNA22) and a 50-nt (DNA50) AT-rich DNA hairpin were investigated using trapped ion mobility spectrometry-mass spectrometry (TIMS-MS) under native starting solvent conditions (e.g., 100 mM aqueous NH4Ac) and collision-induced unfolding/dissociation (CIU/CID) as well as solution fluorescence anisotropy to assess the role of the DNA ligand when binding to the HMGA2 protein. CIU-TIMS-CID-MS/MS experiments showed a significant reduction of the conformational space and charge-state distribution accompanied by an energy stability increase of the native HMGA2 upon DNA binding. Fluorescence anisotropy experiments and CIU-TIMS-CID-MS/MS demonstrated for the first time that HMGA2 binds with high affinity to the minor groove of AT-rich DNA oligomers and with lower affinity to the major groove of AT-rich DNA oligomers (minor groove occupied by a minor groove binder Hoechst 33258). The HMGA2·DNA22 complex (18.2 kDa) 1:1 and 1:2 stoichiometry suggests that two of the AT-hook sites are accessible for DNA binding, while the other AT-hook site is probably coordinated by the C-terminal motif peptide (CTMP). The HMGA2 transition from disordered to ordered upon DNA binding is driven by the interaction of the three basic AT-hook residues with the minor and/or major grooves of AT-rich DNA oligomers.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Zhou X; Benson KF; Ashar HR; Chada K Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 1995, 376 (6543), 771–774. - PubMed
-
- Chung J; Zhang X; Collins B; Sper RB; Gleason K; Simpson S; Koh S; Sommer J; Flowers WL; Petters RM; Piedrahita JA High mobility group A2 (HMGA2) deficiency in pigs leads to dwarfism, abnormal fetal resource allocation, and cryptorchidism. Proc. Natl. Acad. Sci. U.S.A 2018, 115 (21), 5420–5425. - PMC - PubMed
-
- Weedon MN; Lettre G; Freathy RM; Lindgren CM; Voight BF; Perry JR; Elliott KS; Hackett R; Guiducci C; Shields B; Zeggini E; Lango H; Lyssenko V; Timpson NJ; Burtt NP; Rayner NW; Saxena R; Ardlie K; Tobias JH; Ness AR; Ring SM; Palmer CN; Morris AD; Peltonen L; Salomaa V; Diabetes Genetics; Davey Smith G; Wellcome Trust Case Control; Groop, L. C.; Hattersley AT; McCarthy MI; Hirschhorn JN; Frayling TM A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat. Genet 2007, 39 (10), 1245–1250. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

