Sex-specific variation in R-loop formation in Drosophila melanogaster
- PMID: 35687614
- PMCID: PMC9223372
- DOI: 10.1371/journal.pgen.1010268
Sex-specific variation in R-loop formation in Drosophila melanogaster
Abstract
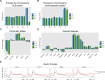
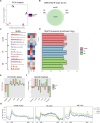
R-loops are three-stranded nucleotide structures consisting of a DNA:RNA hybrid and a displaced ssDNA non-template strand. Previous work suggests that R-loop formation is primarily determined by the thermodynamics of DNA:RNA binding, which are governed by base composition (e.g., GC skew) and transcription-induced DNA superhelicity. However, R-loops have been described at genomic locations that lack these properties, suggesting that they may serve other context-specific roles. To better understand the genetic determinants of R-loop formation, we have characterized the Drosophila melanogaster R-loop landscape across strains and between sexes using DNA:RNA immunoprecipitation followed by high-throughput sequencing (DRIP-seq). We find that R-loops are associated with sequence motifs that are G-rich or exhibit G/C skew, as well as highly expressed genes, tRNAs, and small nuclear RNAs, consistent with a role for DNA sequence and torsion in R-loop specification. However, we also find motifs associated with R-loops that are A/T-rich and lack G/C skew as well as a subset of R-loops that are enriched in polycomb-repressed chromatin. Differential enrichment analysis reveals a small number of sex-biased R-loops: while non-differentially enriched and male-enriched R-loops form at similar genetic features and chromatin states and contain similar sequence motifs, female-enriched R-loops form at unique genetic features, chromatin states, and sequence motifs and are associated with genes that show ovary-biased expression. Male-enriched R-loops are most abundant on the dosage-compensated X chromosome, where R-loops appear stronger compared to autosomal R-loops. R-loop-containing genes on the X chromosome are dosage-compensated yet show lower MOF binding and reduced H4K16ac compared to R-loop-absent genes, suggesting that H4K16ac or MOF may attenuate R-loop formation. Collectively, these results suggest that R-loop formation in vivo is not fully explained by DNA sequence and topology and raise the possibility that a distinct subset of these hybrid structures plays an important role in the establishment and maintenance of epigenetic differences between sexes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Complex determinants of R-loop formation at transposable elements and major DNA satellites.Genetics. 2025 Apr 17;229(4):iyaf035. doi: 10.1093/genetics/iyaf035. Genetics. 2025. PMID: 40036798 Free PMC article.
-
R-loop Mapping and Characterization During Drosophila Embryogenesis Reveals Developmental Plasticity in R-loop Signatures.J Mol Biol. 2022 Jul 15;434(13):167645. doi: 10.1016/j.jmb.2022.167645. Epub 2022 May 21. J Mol Biol. 2022. PMID: 35609632 Free PMC article.
-
Characterization of R-Loop Structures Using Single-Molecule R-Loop Footprinting and Sequencing.Methods Mol Biol. 2020;2161:209-228. doi: 10.1007/978-1-0716-0680-3_15. Methods Mol Biol. 2020. PMID: 32681515 Free PMC article.
-
Building an integrated view of R-loops, transcription, and chromatin.DNA Repair (Amst). 2025 May;149:103832. doi: 10.1016/j.dnarep.2025.103832. Epub 2025 Apr 8. DNA Repair (Amst). 2025. PMID: 40222192 Review.
-
Emerging roles for R-loop structures in the management of topological stress.J Biol Chem. 2020 Apr 3;295(14):4684-4695. doi: 10.1074/jbc.REV119.006364. Epub 2020 Feb 27. J Biol Chem. 2020. PMID: 32107311 Free PMC article. Review.
Cited by
-
Single-cell RNA-seq of Drosophila miranda testis reveals the evolution and trajectory of germline sex chromosome regulation.PLoS Biol. 2024 Apr 30;22(4):e3002605. doi: 10.1371/journal.pbio.3002605. eCollection 2024 Apr. PLoS Biol. 2024. PMID: 38687805 Free PMC article.
-
Complex determinants of R-loop formation at transposable elements and major DNA satellites.Genetics. 2025 Apr 17;229(4):iyaf035. doi: 10.1093/genetics/iyaf035. Genetics. 2025. PMID: 40036798 Free PMC article.
References
-
- Bersani F, Lee E, Kharchenko PV, Xu AW, Liu M, Xega K, et al.. Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proc Natl Acad Sci U S A. 2015;112(49):15148–53. Epub 20151102. doi: 10.1073/pnas.1518008112 ; PubMed Central PMCID: PMC4679016. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

