Follicular Lymphoma Microenvironment Characteristics Associated with Tumor Cell Mutations and MHC Class II Expression
- PMID: 35687817
- PMCID: PMC9894575
- DOI: 10.1158/2643-3230.BCD-21-0075
Follicular Lymphoma Microenvironment Characteristics Associated with Tumor Cell Mutations and MHC Class II Expression
Abstract
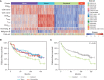
Follicular lymphoma (FL) is a B-cell malignancy with a complex tumor microenvironment that is rich in nonmalignant immune cells. We applied single-cell RNA sequencing to characterize the diverse tumor and immune cell populations of FL and identified major phenotypic subsets of FL T cells, including a cytotoxic CD4 T-cell population. We characterized four major FL subtypes with differential representation or relative depletion of distinct T-cell subsets. By integrating exome sequencing, we observed that somatic mutations are associated with, but not definitive for, reduced MHC expression on FL cells. In turn, expression of MHCII genes by FL cells was associated with significant differences in the proportions and targetable immunophenotypic characteristics of T cells. This provides a classification framework of the FL microenvironment in association with FL genotypes and MHC expression, and informs different potential immunotherapeutic strategies based upon tumor cell MHCII expression.
Significance: We have characterized the FL-infiltrating T cells, identified cytotoxic CD4 T cells as an important component that is associated with tumor cell-intrinsic characteristics, and identified sets of targetable immune checkpoints on T cells that differed from FLs with normal versus low MHC expression. See related commentary by Melnick, p. 374. This article is highlighted in the In This Issue feature, p. 369.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures






Comment in
-
Tee-ing up a New Follicular Lymphoma Classification System.Blood Cancer Discov. 2022 Sep 6;3(5):374-377. doi: 10.1158/2643-3230.BCD-22-0090. Blood Cancer Discov. 2022. PMID: 35917283 Free PMC article.
Comment on
-
Tee-ing up a New Follicular Lymphoma Classification System.Blood Cancer Discov. 2022 Sep 6;3(5):374-377. doi: 10.1158/2643-3230.BCD-22-0090. Blood Cancer Discov. 2022. PMID: 35917283 Free PMC article.
References
-
- Green MR, Alizadeh AA. Common progenitor cells in mature B-cell malignancies: implications for therapy. Curr Opin Hematol 2014;21:333–40. - PubMed
-
- Ennishi D, Takata K, Beguelin W, Duns G, Mottok A, Farinha P, et al. . Molecular and genetic characterization of MHC deficiency identifies EZH2 as therapeutic target for enhancing immune recognition. Cancer Discov 2019;9:546–63. - PubMed
-
- Milpied P, Cervera-Marzal I, Mollichella ML, Tesson B, Brisou G, Traverse-Glehen A, et al. . Human germinal center transcriptional programs are de-synchronized in B cell lymphoma. Nat Immunol 2018;19:1013–24. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

