Loss of MMR and TGFBR2 Increases the Susceptibility to Microbiota-Dependent Inflammation-Associated Colon Cancer
- PMID: 35688320
- PMCID: PMC9421583
- DOI: 10.1016/j.jcmgh.2022.05.010
Loss of MMR and TGFBR2 Increases the Susceptibility to Microbiota-Dependent Inflammation-Associated Colon Cancer
Abstract
Background and aims: Mutations in DNA mismatch repair (MMR) genes are causative in Lynch syndrome and a significant proportion of sporadic colorectal cancers (CRCs). MMR-deficient (dMMR) CRCs display increased mutation rates, with mutations frequently accumulating at short repetitive DNA sequences throughout the genome (microsatellite instability). The TGFBR2 gene is one of the most frequently mutated genes in dMMR CRCs. Therefore, we generated an animal model to study how the loss of both TGFBR2 signaling impacts dMMR-driven intestinal tumorigenesis in vivo and explore the impact of the gut microbiota.
Methods: We generated VCMsh2/Tgfbr2 mice in which Msh2loxP and Tgfbr2loxP alleles are inactivated by Villin-Cre recombinase in the intestinal epithelium. VCMsh2/Tgfbr2 mice were analyzed for their rate of intestinal cancer development and for the mutational spectra and gene expression profiles of tumors. In addition, we assessed the impact of chemically induced chronic inflammation and gut microbiota composition on colorectal tumorigenesis.
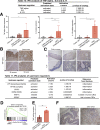
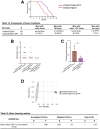
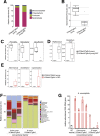
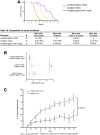
Results: VCMsh2/Tgfbr2 mice developed small intestinal adenocarcinomas and CRCs with histopathological features highly similar to CRCs in Lynch syndrome patients. The CRCs in VCMsh2/Tgfbr2 mice were associated with the presence of colitis and displayed genetic and histological features that resembled inflammation-associated CRCs in human patients. The development of CRCs in VCMsh2/Tgfbr2 mice was strongly modulated by the gut microbiota composition, which in turn was impacted by the TGFBR2 status of the tumors.
Conclusions: Our results demonstrate a synergistic interaction between MMR and TGFBR2 inactivation in inflammation-associated colon tumorigenesis and highlight the crucial impact of the gut microbiota on modulating the incidence of inflammation-associated CRCs.
Keywords: Colon Cancer; DNA Mismatch Repair; Gut Microbiota; Inflammation.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Figures









References
-
- Keum N., Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16:713–732. - PubMed
-
- Barrett M., Hand C.K., Shanahan F., Murphy T., O'Toole P.W. Mutagenesis by microbe: the role of the microbiota in shaping the cancer genome. Trends Cancer. 2020;6:277–287. - PubMed
-
- Janney A., Powrie F., Mann E.H. Host-microbiota maladaptation in colorectal cancer. Nature. 2020;585:509–517. - PubMed
-
- Gupta D., Heinen C.D. The mismatch repair-dependent DNA damage response: Mechanisms and implications. DNA Repair (Amst) 2019;78:60–69. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

