Linear programming based gene expression model (LPM-GEM) predicts the carbon source for Bacillus subtilis
- PMID: 35689204
- PMCID: PMC9188260
- DOI: 10.1186/s12859-022-04742-7
Linear programming based gene expression model (LPM-GEM) predicts the carbon source for Bacillus subtilis
Abstract
Background: Elucidating cellular metabolism led to many breakthroughs in biotechnology, synthetic biology, and health sciences. To date, deriving metabolic fluxes by 13C tracer experiments is the most prominent approach for studying metabolic fluxes quantitatively, often with high accuracy and precision. However, the technique has a high demand for experimental resources. Alternatively, flux balance analysis (FBA) has been employed to estimate metabolic fluxes without labeling experiments. It is less informative but can benefit from the low costs and low experimental efforts and gain flux estimates in experimentally difficult conditions. Methods to integrate relevant experimental data have been emerged to improve FBA flux estimations. Data from transcription profiling is often selected since it is easy to generate at the genome scale, typically embedded by a discretization of differential and non-differential expressed genes coding for the respective enzymes.
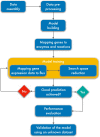
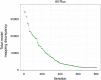
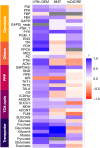
Result: We established the novel method Linear Programming based Gene Expression Model (LPM-GEM). LPM-GEM linearly embeds gene expression into FBA constraints. We implemented three strategies to reduce thermodynamically infeasible loops, which is a necessary prerequisite for such an omics-based model building. As a case study, we built a model of B. subtilis grown in eight different carbon sources. We obtained good flux predictions based on the respective transcription profiles when validating with 13C tracer based metabolic flux data of the same conditions. We could well predict the specific carbon sources. When testing the model on another, unseen dataset that was not used during training, good prediction performance was also observed. Furthermore, LPM-GEM outperformed a well-established model building methods.
Conclusion: Employing LPM-GEM integrates gene expression data efficiently. The method supports gene expression-based FBA models and can be applied as an alternative to estimate metabolic fluxes when tracer experiments are inappropriate.
Keywords: Bacillus subtilis; Carbon source; Constraint-based modeling; Flux balance analysis; Mixed-integer linear programming; Thermodynamically infeasible loops; Transcriptomics.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Chiewchankaset P, Siriwat W, Suksangpanomrung M, Boonseng O, Meechai A, Tanticharoen M, Kalapanulak S, Saithong T. Understanding carbon utilization routes between high and low starch-producing cultivars of cassava through Flux Balance Analysis. Sci Rep. 2019;9(1):2964. doi: 10.1038/s41598-019-39920-w. - DOI - PMC - PubMed
-
- Lu H, Liu X, Huang M, Xia J, Chu J, Zhuang Y, Zhang S, Noorman H. Integrated isotope-assisted metabolomics and (13)C metabolic flux analysis reveals metabolic flux redistribution for high glucoamylase production by Aspergillus niger. Microb Cell Fact. 2015;14:147. doi: 10.1186/s12934-015-0329-y. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

