CRISPR-Cas, Argonaute proteins and the emerging landscape of amplification-free diagnostics
- PMID: 35690249
- PMCID: PMC9181078
- DOI: 10.1016/j.ymeth.2022.06.002
CRISPR-Cas, Argonaute proteins and the emerging landscape of amplification-free diagnostics
Abstract
Polymerase Chain Reaction (PCR) is the reigning gold standard for molecular diagnostics. However, the SARS-CoV-2 pandemic reveals an urgent need for new diagnostics that provide users with immediate results without complex procedures or sophisticated equipment. These new demands have stimulated a tsunami of innovations that improve turnaround times without compromising the specificity and sensitivity that has established PCR as the paragon of diagnostics. Here we briefly introduce the origins of PCR and isothermal amplification, before turning to the emergence of CRISPR-Cas and Argonaute proteins, which are being coupled to fluorimeters, spectrometers, microfluidic devices, field-effect transistors, and amperometric biosensors, for a new generation of nucleic acid-based diagnostics.
Keywords: Argonaute; CRISPR-Cas; Molecular diagnostics; Nucleic acid detection; TnpB/IscB.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: B.W. is the founder of SurGene and VIRIS Detection Systems. B.W., A.S.-F., A. Nemudraia, and A. Nemudryi are inventors on patent applications related to CRISPR-Cas systems and applications thereof.
Figures

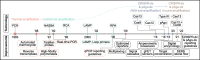



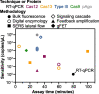
References
-
- Mullis K.B. The Polymerase Chain Reaction (Nobel Lecture) Angew. Chemie Int. Ed. English. 1994;33:1209–1213. doi: 10.1002/anie.199412091. - DOI
-
- K.B. Mullis, H.A. Erlich, N. Arnheim, G.T. Horn, R.K. Saiki, S.J. Scharf. Process for amplifying, detecting, and/or-cloning nucleic acid sequences. 828144, 1987.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

