Muscle regeneration affects Adeno Associated Virus 1 mediated transgene transcription
- PMID: 35690627
- PMCID: PMC9188557
- DOI: 10.1038/s41598-022-13405-9
Muscle regeneration affects Adeno Associated Virus 1 mediated transgene transcription
Abstract
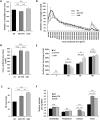
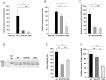
Duchenne muscular dystrophy is a severe neuromuscular disease causing a progressive muscle wasting due to mutations in the DMD gene that lead to the absence of dystrophin protein. Adeno-associated virus (AAV)-based therapies aiming to restore dystrophin in muscles, by either exon skipping or microdystrophin expression, are very promising. However, the absence of dystrophin induces cellular perturbations that hinder AAV therapy efficiency. We focused here on the impact of the necrosis-regeneration process leading to nuclear centralization in myofiber, a common feature of human myopathies, on AAV transduction efficiency. We generated centronucleated myofibers by cardiotoxin injection in wild-type muscles prior to AAV injection. Intramuscular injections of AAV1 vectors show that transgene expression was drastically reduced in regenerated muscles, even when the AAV injection occurred 10 months post-regeneration. We show also that AAV genomes were not lost from cardiotoxin regenerated muscle and were properly localised in the myofiber nuclei but were less transcribed leading to muscle transduction defect. A similar defect was observed in muscles of the DMD mouse model mdx. Therefore, the regeneration process per se could participate to the AAV-mediated transduction defect observed in dystrophic muscles which may limit AAV-based therapies.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

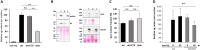

References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

