A single hepatitis B virus genome with a reporter allows the entire viral life cycle to be monitored in primary human hepatocytes
- PMID: 35691027
- PMCID: PMC9426382
- DOI: 10.1002/hep4.2018
A single hepatitis B virus genome with a reporter allows the entire viral life cycle to be monitored in primary human hepatocytes
Abstract
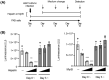
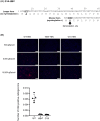
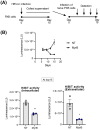
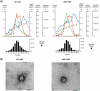
For the development of antiviral agents to eliminate hepatitis B virus (HBV), it is essential to establish an HBV cell culture system that can easily monitor HBV infection. Here, we created a novel HBV infection monitoring system using a luminescent 11-amino acid reporter, the high-affinity subunit of nano-luciferase binary technology (HiBiT). The HiBiT-coding sequence was inserted at the N-terminus of preS1 in a 1.2-fold plasmid encoding a genotype C HBV genome. After transfection of HepG2 cells with this HiBiT-containing plasmid, the supernatant was used to prepare a recombinant cell culture-derived virus (HiBiT-HBVcc). Primary human hepatocytes (PXB) were inoculated with HiBiT-HBVcc. Following inoculation, intracellular and extracellular HiBiT activity and the levels of various HBV markers were determined. Reinfection of naive PXB cells with HiBiT-HBVcc prepared from HiBiT-HBVcc-infected PXB cells was analyzed. When PXB cells were infected with HiBiT-HBVcc at several titers, extracellular HiBiT activity was detected in a viral titer-dependent manner and was correlated with intracellular HiBiT activity. Inhibitors of HBV entry or replication suppressed extracellular HiBiT activity. Viral DNA, RNA, and proteins were detectable, including covalently closed circular DNA, by Southern blot analysis. The synthesis of relaxed-circular DNA from single-stranded DNA in HiBiT-HBV decreased to one third of that of wild-type HBV, and the infectivity of HiBiT-HBVcc decreased to one tenth of that of wild-type HBVcc. HiBiT-HBVcc prepared from PXB cells harboring HiBiT-HBV was able to infect naive PXB cells. Conclusions: Recombinant HiBiT-HBV can undergo the entire viral life cycle, thus facilitating high-throughput screening for HBV infection in vitro using supernatants. This system will be a powerful tool for developing antiviral agents.
© 2022 The Authors. Hepatology Communications published by Wiley Periodicals LLC on behalf of American Association for the Study of Liver Diseases.
Conflict of interest statement
Nothing to report.
Figures



References
-
- Polaris Observatory Collaborators . Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol. 2018;3:383–403. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

