Methyltransferases: Functions and Applications
- PMID: 35691829
- PMCID: PMC9539859
- DOI: 10.1002/cbic.202200212
Methyltransferases: Functions and Applications
Abstract
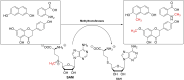
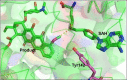
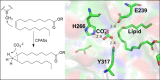
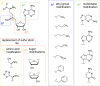
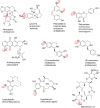
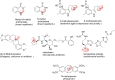
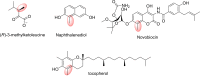
In this review the current state-of-the-art of S-adenosylmethionine (SAM)-dependent methyltransferases and SAM are evaluated. Their structural classification and diversity is introduced and key mechanistic aspects presented which are then detailed further. Then, catalytic SAM as a target for drugs, and approaches to utilise SAM as a cofactor in synthesis are introduced with different supply and regeneration approaches evaluated. The use of SAM analogues are also described. Finally O-, N-, C- and S-MTs, their synthetic applications and potential for compound diversification is given.
Keywords: S-adenosyl-l-methionine; biocatalysis; enzymes; methyltransferases.
© 2022 The Authors. ChemBioChem published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



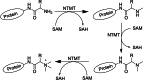
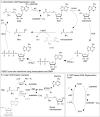
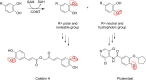
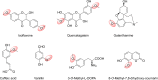
References
-
- Constable D. J. C., Dunn P. J., Hayler J. D., Humphrey G. R., J. L. Leazer Jr. , Linderman R. J., Lorenz K., Manley J., Pearlman B. A., Wells A., Zaks A., Zhang T. Y., Green Chem. 2007, 9, 411–420.
-
- None
-
- Richter M., Nat. Prod. Rep. 2013, 30, 1324–1345; - PubMed
-
- Chen H., Wang Z., Cai H., Zhou C., World J. Microbiol. Biotechnol. 2016, 32, 153; - PubMed
-
- Cantoni G. L., Annu. Rev. Biochem. 1975, 44, 435–451. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

