Identification of Immune-Related Genes for Risk Stratification in Multiple Myeloma Based on Whole Bone Marrow Gene Expression Profiling
- PMID: 35692836
- PMCID: PMC9178200
- DOI: 10.3389/fgene.2022.897886
Identification of Immune-Related Genes for Risk Stratification in Multiple Myeloma Based on Whole Bone Marrow Gene Expression Profiling
Abstract
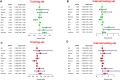
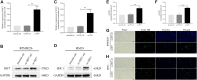
Background: Multiple myeloma (MM) is characterized by abnormal proliferation of bone marrow clonal plasma cells. Tumor immunotherapy, a new therapy that has emerged in recent years, offers hope to patients, and studying the expression characteristics of immune-related genes (IRGs) based on whole bone marrow gene expression profiling (GEP) in MM patients can help guide personalized immunotherapy. Methods: In this study, we explored the potential prognostic value of IRGs in MM by combining GEP and clinical data from the GEO database. We identified hub IRGs and transcription factors (TFs) associated with disease progression by Weighted Gene Co-expression Network Analysis (WGCNA), and modeled immune-related prognostic signature by univariate and multivariate Cox and least absolute shrinkage and selection operator (LASSO) regression analysis. Subsequently, the prognostic ability of signature was verified by multiple statistical methods. Moreover, ssGSEA and GSEA algorithm reveled different immunological characteristics and biological function variation in different risk groups. We mapped the hub IRGs by protein-protein interaction network (PPI) and extracted the top 10 ranked genes. Finally, we conducted vitro assays on two alternative IRGs. Results: Our study identified a total of 14 TFs and 88 IRGs associated with International Staging System (ISS). Ten IRGs were identified by Cox -LASSO regression analysis, and used to develop optimal prognostic signature for overall survival (OS) in MM patients. The 10-IRGs were BDNF, CETP, CD70, LMBR, LTBP1, NENF, NR1D1, NR1H2, PTK2B and SEMA4. In different groups, risk signatures showed excellent survival prediction ability, and MM patients also could be stratified at survival risk. In addition, IRF7 and SHC1 were hub IRGs in PPI network, and the vitro assays proved that they could promote tumor progression. Notably, ssGSEA and GSEA results confirmed that different risk groups could accurately indicate the status of tumor microenvironment (TME) and activation of biological pathways. Conclusion: Our study suggested that immune-related signature could be used as prognostic markers in MM patients.
Keywords: IRF7; SHC1; immune-related genes; multiple myeloma; prognostic model; whole bone marrow sequencing.
Copyright © 2022 Wang, Shi, Meng, Chen and Hou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

