Family Members Additively Repress the Ectopic Expression of BASIC PENTACYSTEINE3 to Prevent Disorders in Arabidopsis Circadian Vegetative Development
- PMID: 35693178
- PMCID: PMC9182635
- DOI: 10.3389/fpls.2022.919946
Family Members Additively Repress the Ectopic Expression of BASIC PENTACYSTEINE3 to Prevent Disorders in Arabidopsis Circadian Vegetative Development
Abstract
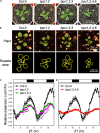
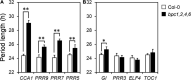
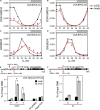
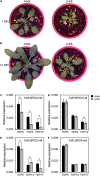
BARLEY B-RECOMBINANT/BASIC PENTACYSTEINE (BBR/BPC) family members are plant-specific GAGA-motif binding factors (GAFs) controlling multiple developmental processes of growth and propagation. BPCs recruit histone remodeling factors for transcriptional repression of downstream targets. It has been revealed that BPCs have an overlapping and antagonistic relationship in regulating development. In this study, we showed disturbances interfering with the homeostasis of BPC expressions impede growth and development. The ectopic expression of BPC3 results in the daily growth defect shown by higher-order bpc mutants. Oscillations of multiple circadian clock genes are phase-delayed in the quadruple mutant of bpc1 bpc2 bpc4 bpc6 (bpc1,2,4,6). By introducing the overexpression of BPC3 into wild-type Arabidopsis, we found that BPC3 is a repressor participating in its repression and repressing multiple regulators essential to the circadian clock. However, the induction of BPC3 overexpression did not fully replicate clock defects shown by the quadruple mutant, indicating that in addition to the BPC3 antagonization, BPC members also cofunction in the circadian clock regulation. A leaf edge defect similar to that shown by bpc1,2,4,6 is also observed under BPC3 induction, accompanied by repression of a subset of TCPs required for the edge formation. This proves that BPC3 is a repressor that must be confined during the vegetative phase. Our findings demonstrate that BPCs form a meticulous repressor network for restricting their repressive functions to molecular mechanisms controlling plant growth and development.
Keywords: Arabidopsis thaliana; BPC transcription factor; TCP transcription factor; circadian clock; leaf development.
Copyright © 2022 Lee, Tsai, Huang and Tsai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Chang S., Puryear J., Cairney J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11 113–116. 10.1385/MB:19:2:201 - DOI
-
- Engelmann W., Johnsson A. (1998). “Rhythms in organ movement,” in Biological Rhythms and Photoperiodism in Plants, eds Lumsden P. J., Millar A. J. (Oxford: BIOS Scientific Publishers; ), 35–50.
-
- Fowler S., Lee K., Onouchi H., Samach A., Richardson K., Morris B., et al. (1999). GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18, 4679–4688. 10.1093/emboj/18.17.4679 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

