A Machine-Learning Approach to Developing a Predictive Signature Based on Transcriptome Profiling of Ground-Glass Opacities for Accurate Classification and Exploring the Immune Microenvironment of Early-Stage LUAD
- PMID: 35693786
- PMCID: PMC9178173
- DOI: 10.3389/fimmu.2022.872387
A Machine-Learning Approach to Developing a Predictive Signature Based on Transcriptome Profiling of Ground-Glass Opacities for Accurate Classification and Exploring the Immune Microenvironment of Early-Stage LUAD
Abstract
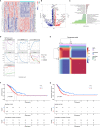
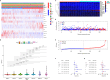
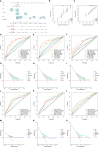
Screening for early-stage lung cancer with low-dose computed tomography is recommended for high-risk populations; consequently, the incidence of pure ground-glass opacity (pGGO) is increasing. Ground-glass opacity (GGO) is considered the appearance of early lung cancer, and there remains an unmet clinical need to understand the pathology of small GGO (<1 cm in diameter). The objective of this study was to use the transcriptome profiling of pGGO specimens <1 cm in diameter to construct a pGGO-related gene risk signature to predict the prognosis of early-stage lung adenocarcinoma (LUAD) and explore the immune microenvironment of GGO. pGGO-related differentially expressed genes (DEGs) were screened to identify prognostic marker genes with two machine learning algorithms. A 15-gene risk signature was constructed from the DEGs that were shared between the algorithms. Risk scores were calculated using the regression coefficients for the pGGO-related DEGs. Patients with Stage I/II LUAD or Stage IA LUAD and high-risk scores had a worse prognosis than patients with low-risk scores. The prognosis of high-risk patients with Stage IA LUAD was almost identical to that of patients with Stage II LUAD, suggesting that treatment strategies for patients with Stage II LUAD may be beneficial in high-risk patients with Stage IA LUAD. pGGO-related DEGs were mainly enriched in immune-related pathways. Patients with high-risk scores and high tumor mutation burden had a worse prognosis and may benefit from immunotherapy. A nomogram was constructed to facilitate the clinical application of the 15-gene risk signature. Receiver operating characteristic curves and decision curve analysis validated the predictive ability of the nomogram in patients with Stage I LUAD in the TCGA-LUAD cohort and GEO datasets.
Keywords: GEO; GGO (ground-glass opacity); LUAD; TCGA; prognosis.
Copyright © 2022 Zhao, Yin, Peng, Cai, He, Shi, Peng, Tu, Li, Li, Tao, Peng, Wang and Yu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Hattori A, Hirayama S, Matsunaga T, Hayashi T, Takamochi K, Oh S, et al. Distinct Clinicopathologic Characteristics and Prognosis Based on the Presence of Ground Glass Opacity Component in Clinical Stage IA Lung Adenocarcinoma. J Thorac Oncol (2019) 14(2):265–75. doi: 10.1016/j.jtho.2018.09.026 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

