Electroacupuncture Ameliorates Mechanical Allodynia of a Rat Model of CRPS-I via Suppressing NLRP3 Inflammasome Activation in Spinal Cord Dorsal Horn Neurons
- PMID: 35693886
- PMCID: PMC9174662
- DOI: 10.3389/fncel.2022.826777
Electroacupuncture Ameliorates Mechanical Allodynia of a Rat Model of CRPS-I via Suppressing NLRP3 Inflammasome Activation in Spinal Cord Dorsal Horn Neurons
Abstract
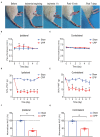
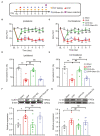
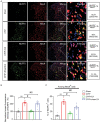
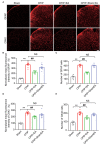
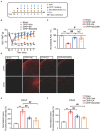
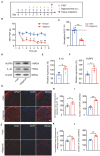
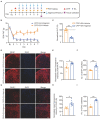
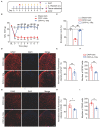
Complex regional pain syndrome type-I (CRPS-I) is a chronic neurological disorder that results in severe pain and affects patients' life quality. Conventional therapies usually lack effectiveness. Electroacupuncture (EA) is an effective physical therapy for relieving CRPS-I pain. However, the mechanism underlying EA-induced analgesia on CRPS-I still remain unknown. Spinal NLRP3 inflammasome was recently identified to contribute to pain and neuroinflammation in a rat model of CRPS-I by our group. Here, we aimed to study whether EA could inhibit spinal NLRP3 inflammasome activation, thus resulting in pain relief and attenuation of spinal neuroinflammation in the rat model of CRPS-I. We established the rat chronic post-ischemic pain (CPIP) model to mimic CRPS-I. CPIP rats developed remarkable mechanical allodynia that could be relieved by daily EA intervention. NLRP3 inflammasome was activated in spinal cord dorsal horn (SCDH) of CPIP rats, accompanied with over-production of pro-inflammatory cytokine IL-1β. Immunostaining revealed that the cellular distribution of NLRP3 was predominantly located in SCDH neurons. Pharmacological activation of NLRP3 inflammasome per se is sufficient to produce persistent mechanical allodynia in naïve animals, whereas blocking NLRP3 inflammasome attenuates mechanical allodynia of CPIP rats. EA exclusively reduced NLRP3 overexpression in SCDH neurons and attenuated spinal glial cell over-activation in CPIP rats. EA-induced anti-allodynia with attenuation of spinal glial cell over-activation were all mimicked by intrathecal blocking NLRP3 inflammasome and reversed by activating NLRP3 inflammasome, respectively, through pharmacological methods. Finally, spinal blocking IL-1β attenuated mechanical allodynia and spinal glial cell over-activation in CPIP rats, resembling the effects of EA. In all, these results demonstrate that spinal NLRP3 inflammasome activation contributes to mechanical allodynia of the rat model of CRPS-I and EA ameliorates mechanical allodynia through inhibiting NLRP3 inflammasome activation in SCDH neurons. Our study further supports EA can be used as an effective treatment for CRPS-I.
Keywords: allodynia; complex regional pain syndrome; electroacupuncture; glial cell; inflammasome; spinal cord.
Copyright © 2022 Zhang, Chen, Hu, Wang, Nie, Yin, Li, Wei, Liu, Tai, Fang, Shao, Jin, Fang and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Electroacupuncture Alleviates Mechanical Allodynia in a Rat Model of Complex Regional Pain Syndrome Type-I via Suppressing Spinal CXCL12/CXCR4 Signaling.J Pain. 2020 Sep-Oct;21(9-10):1060-1074. doi: 10.1016/j.jpain.2020.01.007. Epub 2020 Jun 25. J Pain. 2020. PMID: 32006698
-
Expression profiling of spinal cord dorsal horn in a rat model of complex regional pain syndrome type-I uncovers potential mechanisms mediating pain and neuroinflammation responses.J Neuroinflammation. 2020 May 23;17(1):162. doi: 10.1186/s12974-020-01834-0. J Neuroinflammation. 2020. PMID: 32446302 Free PMC article.
-
CXCL13 contributes to chronic pain of a mouse model of CRPS-I via CXCR5-mediated NF-κB activation and pro-inflammatory cytokine production in spinal cord dorsal horn.J Neuroinflammation. 2023 May 8;20(1):109. doi: 10.1186/s12974-023-02778-x. J Neuroinflammation. 2023. PMID: 37158939 Free PMC article.
-
Ion Channels and G-Protein-Coupled Receptors Involved in the Development of Chronic Post-ischemic Pain (CPIP): A Model of Complex Regional Pain Syndrome (CRPS-I).Mol Neurobiol. 2025 May 19. doi: 10.1007/s12035-025-05043-9. Online ahead of print. Mol Neurobiol. 2025. PMID: 40383737 Review.
-
Chronic post-ischemic pain (CPIP) a model of complex regional pain syndrome (CRPS-I): Role of oxidative stress and inflammation.Biochem Pharmacol. 2024 Nov;229:116506. doi: 10.1016/j.bcp.2024.116506. Epub 2024 Aug 23. Biochem Pharmacol. 2024. PMID: 39182734 Review.
Cited by
-
Electroacupuncture improves gout arthritis pain via attenuating ROS-mediated NLRP3 inflammasome overactivation.Chin Med. 2023 Jul 18;18(1):86. doi: 10.1186/s13020-023-00800-1. Chin Med. 2023. PMID: 37464384 Free PMC article.
-
Chemokine CXCL13-CXCR5 signaling in neuroinflammation and pathogenesis of chronic pain and neurological diseases.Cell Mol Biol Lett. 2024 Oct 29;29(1):134. doi: 10.1186/s11658-024-00653-y. Cell Mol Biol Lett. 2024. PMID: 39472796 Free PMC article. Review.
-
Immune mechanisms in vulvodynia: key roles for mast cells and fibroblasts.Front Cell Infect Microbiol. 2023 Jun 8;13:1215380. doi: 10.3389/fcimb.2023.1215380. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37360527 Free PMC article. Review.
-
Electroacupuncture Alleviates Pain by Suppressing P2Y12R-Dependent Microglial Activation in Monoarthritic Rats.Neurochem Res. 2024 May;49(5):1268-1277. doi: 10.1007/s11064-024-04114-y. Epub 2024 Feb 10. Neurochem Res. 2024. PMID: 38337134
-
Research trends of acupuncture therapy for painful peripheral nervous system diseases from 2004 to 2023: a bibliometric and meta-analysis.Front Neurol. 2025 Mar 14;16:1510331. doi: 10.3389/fneur.2025.1510331. eCollection 2025. Front Neurol. 2025. PMID: 40162011 Free PMC article.
References
LinkOut - more resources
Full Text Sources

