Fast, Simple, and Highly Specific Molecular Detection of Porphyromonas gingivalis Using Isothermal Amplification and Lateral Flow Strip Methods
- PMID: 35694545
- PMCID: PMC9174636
- DOI: 10.3389/fcimb.2022.895261
Fast, Simple, and Highly Specific Molecular Detection of Porphyromonas gingivalis Using Isothermal Amplification and Lateral Flow Strip Methods
Abstract
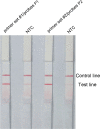
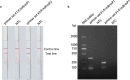
Porphyromonas gingivalis is an important oral pathogen that causes periodontal disease and is difficult to culture under conventional conditions. Therefore, a reliable technique for detecting this pathogenic bacterium is required. Here, isothermal recombinase polymerase amplification (RPA), a new nucleic acid amplification method, was combined with a visualization method based on nanoparticle-based lateral flow strips (LFS) for the rapid detection of P. gingivalis. The species-specific 16S rRNA sequence of P. gingivalis was used as the target for RPA, and a set of specific primer-probe combinations were designed and screened to amplify the target sequences. As a thermostatic amplification method, the RPA reaction, under optimized conditions, takes only 30 min to complete at a constant temperature (37°C). The amplification reaction products can be detected visually by LFS without any need for special equipment. The RPA-LFS method established for the detection of P. gingivalis was shown to be highly specific in distinguishing P. gingivalis from other pathogenic organisms by using 20 clinical isolates of P. gingivalis and 23 common pathogenic microorganisms. Susceptibility measurements and probit regression analysis were performed with gradient dilutions of P. gingivalis genomic DNA. The method was obtained to be highly sensitive, with a detection limit of 9.27 CFU per reaction at 95% probability. By analyzing the gingival sulcus fluid specimens from 130 patients with chronic periodontitis, the results showed that the RPA-LFS method detected 118 positive cases and 12 negative cases of P. gingivalis, and the results obtained were consistent with those of a conventional PCR assay. The RPA-LFS method is an efficient, rapid, and convenient diagnostic method that simplifies the tedious process of detecting P. gingivalis.
Keywords: Porphyromonas gingivalis; isothermal (DNA) amplification; lateral flow strip; periodontal disease; rapid detection.
Copyright © 2022 Ge, Wang, Hu, Wang, Gao and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Alhogail S., Suaifan G., Bizzarro S., Kaman W. E., Bikker F. J., Weber K., et al. (2018). On Site Visual Detection of Porphyromonas Gingivalis Related Periodontitis by Using a Magnetic-Nanobead Based Assay for Gingipains Protease Biomarkers. Mikrochim Acta 185 (2), 149. doi: 10.1007/s00604-018-2677-x - DOI - PubMed
-
- Ambrosio N., Marín M. J., Laguna E., Herrera D., Sanz M., Figuero E. (2019). Detection and Quantification of Porphyromonas Gingivalis and Aggregatibacter Actinomycetemcomitans in Bacteremia Induced by Interdental Brushing in Periodontally Healthy and Periodontitis Patients. Arch. Oral. Biol. 98, 213–219. doi: 10.1016/j.archoralbio.2018.11.025 - DOI - PubMed
-
- Boyer B. P., Ryerson C. C., Reynolds H. S., Zambon J. J., Genco R. J., Snyder B. (1996). Colonization by Actinobacillus Actinomycetemcomitans, Porphyromonas Gingivalis and Prevotella Intermedia in Adult Periodontitis Patients as Detected by the Antibody-Based Evalusite Test. J. Clin. Periodontol 23 (5), 477–484. doi: 10.1111/j.1600-051x.1996.tb00578.x - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

