Genomic architecture and functional unit of mimicry supergene in female limited Batesian mimic Papilio butterflies
- PMID: 35694751
- PMCID: PMC9189499
- DOI: 10.1098/rstb.2021.0198
Genomic architecture and functional unit of mimicry supergene in female limited Batesian mimic Papilio butterflies
Abstract
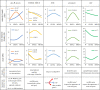
It has long been suggested that dimorphic female-limited Batesian mimicry of two closely related Papilio butterflies, Papilio memnon and Papilio polytes, is controlled by supergenes. Whole-genome sequencing, genome-wide association studies and functional analyses have recently identified mimicry supergenes, including the doublesex (dsx) gene. Although supergenes of both the species are composed of highly divergent regions between mimetic and non-mimetic alleles and are located at the same chromosomal locus, they show critical differences in genomic architecture, particularly with or without an inversion: P. polytes has an inversion, but P. memnon does not. This review introduces and compares the detailed genomic structure of mimicry supergenes in two Papilio species, including gene composition, repetitive sequence composition, breakpoint/boundary site structure, chromosomal inversion and linkage disequilibrium. Expression patterns and functional analyses of the respective genes within or flanking the supergene suggest that dsx and other genes are involved in mimetic traits. In addition, structural comparison of the corresponding region for the mimicry supergene among further Papilio species suggests three scenarios for the evolution of the mimicry supergene between the two Papilio species. The structural features revealed in the Papilio mimicry supergene provide insight into the formation, maintenance and evolution of supergenes. This article is part of the theme issue 'Genomic architecture of supergenes: causes and evolutionary consequences'.
Keywords: Papilio butterflies; chromosomal inversion; female-limited polymorphic mimicry; linkage disequilibrium; supergene; transposon.
Conflict of interest statement
We declare we have no competing interests.
Figures


Similar articles
-
Functional unit of supergene in female-limited Batesian mimicry of Papilio polytes.Genetics. 2023 Feb 9;223(2):iyac177. doi: 10.1093/genetics/iyac177. Genetics. 2023. PMID: 36454671 Free PMC article.
-
Parallel evolution of Batesian mimicry supergene in two Papilio butterflies, P. polytes and P. memnon.Sci Adv. 2018 Apr 18;4(4):eaao5416. doi: 10.1126/sciadv.aao5416. eCollection 2018 Apr. Sci Adv. 2018. PMID: 29675466 Free PMC article.
-
Identification of doublesex alleles associated with the female-limited Batesian mimicry polymorphism in Papilio memnon.Sci Rep. 2016 Oct 6;6:34782. doi: 10.1038/srep34782. Sci Rep. 2016. PMID: 27708422 Free PMC article.
-
The Genomic Architecture and Evolutionary Fates of Supergenes.Genome Biol Evol. 2021 May 7;13(5):evab057. doi: 10.1093/gbe/evab057. Genome Biol Evol. 2021. PMID: 33739390 Free PMC article. Review.
-
The double game of chromosomal inversions in a neotropical butterfly.C R Biol. 2022 May 11;345(1):57-73. doi: 10.5802/crbiol.73. C R Biol. 2022. PMID: 35787620 Review.
Cited by
-
Functional unit of supergene in female-limited Batesian mimicry of Papilio polytes.Genetics. 2023 Feb 9;223(2):iyac177. doi: 10.1093/genetics/iyac177. Genetics. 2023. PMID: 36454671 Free PMC article.
-
Stepwise evolution of a butterfly supergene via duplication and inversion.Philos Trans R Soc Lond B Biol Sci. 2022 Aug;377(1856):20210207. doi: 10.1098/rstb.2021.0207. Epub 2022 Jun 13. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 35694743 Free PMC article.
-
Genomic architecture of supergenes: connecting form and function.Philos Trans R Soc Lond B Biol Sci. 2022 Aug;377(1856):20210192. doi: 10.1098/rstb.2021.0192. Epub 2022 Jun 13. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 35694757 Free PMC article.
-
doublesex Controls Both Hindwing and Abdominal Mimicry Traits in the Female-Limited Batesian Mimicry of Papilio memnon.Front Insect Sci. 2022 Jul 12;2:929518. doi: 10.3389/finsc.2022.929518. eCollection 2022. Front Insect Sci. 2022. PMID: 38468762 Free PMC article.
-
Chromosomal inversions and their impact on insect evolution.Curr Opin Insect Sci. 2024 Dec;66:101280. doi: 10.1016/j.cois.2024.101280. Epub 2024 Oct 5. Curr Opin Insect Sci. 2024. PMID: 39374869 Review.
References
-
- Bates HW. 1862. Contributions to an insect fauna of the Amazon Valley (Lepidoptera: Heliconidae). Trans. Linn. Soc. Lond. 23, 495-556. (10.1111/j.1096-3642.1860.tb00146.x) - DOI
-
- Wallace AR. 1865. On the phenomena of variation and geographical distribution as illustrated by the Papilionidae of the Malayan Region. Trans. Linn. Soc. (Lond.) 25, 1-71. (10.1111/j.1096-3642.1865.tb00178.x) - DOI
-
- Clarke CA, Sheppard PM. 1972. The genetics of the mimetic butterfly Papilio polytes L. Phil. Trans. R. Soc. Lond. B 26, 431-458. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

