Calibration and Validation of a Mechanistic COVID-19 Model for Translational Quantitative Systems Pharmacology - A Proof-of-Concept Model Development for Remdesivir
- PMID: 35694844
- PMCID: PMC9349538
- DOI: 10.1002/cpt.2686
Calibration and Validation of a Mechanistic COVID-19 Model for Translational Quantitative Systems Pharmacology - A Proof-of-Concept Model Development for Remdesivir
Abstract
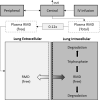
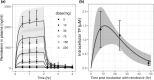
With the ongoing global pandemic of coronavirus disease 2019 (COVID-19), there is an urgent need to accelerate the traditional drug development process. Many studies identified potential COVID-19 therapies based on promising nonclinical data. However, the poor translatability from nonclinical to clinical settings has led to failures of many of these drug candidates in the clinical phase. In this study, we propose a mechanism-based, quantitative framework to translate nonclinical findings to clinical outcome. Adopting a modularized approach, this framework includes an in silico disease model for COVID-19 (virus infection and human immune responses) and a pharmacological component for COVID-19 therapies. The disease model was able to reproduce important longitudinal clinical data for patients with mild and severe COVID-19, including viral titer, key immunological cytokines, antibody responses, and time courses of lymphopenia. Using remdesivir as a proof-of-concept example of model development for the pharmacological component, we developed a pharmacological model that describes the conversion of intravenously administered remdesivir as a prodrug to its active metabolite nucleoside triphosphate through intracellular metabolism and connected it to the COVID-19 disease model. After being calibrated with the placebo arm data, our model was independently and quantitatively able to predict the primary endpoint (time to recovery) of the remdesivir clinical study, Adaptive Covid-19 Clinical Trial (ACTT). Our work demonstrates the possibility of quantitatively predicting clinical outcome based on nonclinical data and mechanistic understanding of the disease and provides a modularized framework to aid in candidate drug selection and clinical trial design for COVID-19 therapeutics.
© 2022 The Authors. Clinical Pharmacology & Therapeutics © 2022 American Society for Clinical Pharmacology and Therapeutics. This article has been contributed to by U.S. Government employees and their work is in the public domain in the USA.
Conflict of interest statement
The authors declared no competing interests for this work. This report is not an official US Food and Drug Administration guidance or policy statement. No official support or endorsement by the US Food and Drug Administration is intended or should be inferred. As an Associate Editor for
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

