Evaluation of Metagenomic and Targeted Next-Generation Sequencing Workflows for Detection of Respiratory Pathogens from Bronchoalveolar Lavage Fluid Specimens
- PMID: 35695488
- PMCID: PMC9297812
- DOI: 10.1128/jcm.00526-22
Evaluation of Metagenomic and Targeted Next-Generation Sequencing Workflows for Detection of Respiratory Pathogens from Bronchoalveolar Lavage Fluid Specimens
Abstract
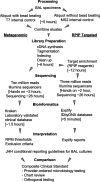
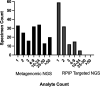
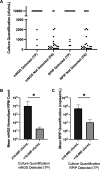
Next-generation sequencing (NGS) workflows applied to bronchoalveolar lavage (BAL) fluid specimens could enhance the detection of respiratory pathogens, although optimal approaches are not defined. This study evaluated the performance of the Respiratory Pathogen ID/AMR (RPIP) kit (Illumina, Inc.) with automated Explify bioinformatic analysis (IDbyDNA, Inc.), a targeted NGS workflow enriching specific pathogen sequences and antimicrobial resistance (AMR) markers, and a complementary untargeted metagenomic workflow with in-house bioinformatic analysis. Compared to a composite clinical standard consisting of provider-ordered microbiology testing, chart review, and orthogonal testing, both workflows demonstrated similar performances. The overall agreement for the RPIP targeted workflow was 65.6% (95% confidence interval, 59.2 to 71.5%), with a positive percent agreement (PPA) of 45.9% (36.8 to 55.2%) and a negative percent agreement (NPA) of 85.7% (78.1 to 91.5%). The overall accuracy for the metagenomic workflow was 67.1% (60.9 to 72.9%), with a PPA of 56.6% (47.3 to 65.5%) and an NPA of 77.2% (68.9 to 84.1%). The approaches revealed pathogens undetected by provider-ordered testing (Ureaplasma parvum, Tropheryma whipplei, severe acute respiratory syndrome coronavirus 2 [SARS-CoV-2], rhinovirus, and cytomegalovirus [CMV]), although not all pathogens detected by provider-ordered testing were identified by the NGS workflows. The RPIP targeted workflow required more time and reagents for library preparation but streamlined bioinformatic analysis, whereas the metagenomic assay was less demanding technically but required complex bioinformatic analysis. The results from both workflows were interpreted utilizing standardized criteria, which is necessary to avoid reporting nonpathogenic organisms. The RPIP targeted workflow identified AMR markers associated with phenotypic resistance in some bacteria but incorrectly identified blaOXA genes in Pseudomonas aeruginosa as being associated with carbapenem resistance. These workflows could serve as adjunctive testing with, but not as a replacement for, standard microbiology techniques.
Keywords: diagnostics; lower respiratory tract infection; next-generation sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Miao Q, Ma Y, Wang Q, Pan J, Zhang Y, Jin W, Yao Y, Su Y, Huang Y, Wang M, Li B, Li H, Zhou C, Li C, Ye M, Xu X, Li Y, Hu B. 2018. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis 67:S231–S240. doi:10.1093/cid/ciy693. - DOI - PubMed
-
- Zhou H, Larkin PMK, Zhao D, Ma Q, Yao Y, Wu X, Wang J, Zhou X, Li Y, Wang G, Feng M, Wu L, Chen J, Zhou C, Hua X, Zhou J, Yang S, Yu Y. 2021. Clinical impact of metagenomic next-generation sequencing of bronchoalveolar lavage in the diagnosis and management of pneumonia: a multicenter prospective observational study. J Mol Diagn 23:1259–1268. doi:10.1016/j.jmoldx.2021.06.007. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

