Fecal carriage of vanB antibiotic resistance gene affects adipose tissue function under vancomycin use
- PMID: 35695620
- PMCID: PMC9196849
- DOI: 10.1080/19490976.2022.2083905
Fecal carriage of vanB antibiotic resistance gene affects adipose tissue function under vancomycin use
Abstract
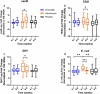
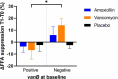
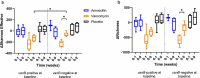
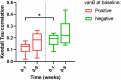
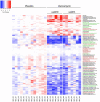
Detrimental consequences of antibiotic treatment may include long-lasting disruption of the gut microbiota. Previous studies found no negative effects of antibiotics on metabolic health, although individualized responses were observed. Here, we aimed to investigate the subject-specific response to vancomycin use in tissue-specific insulin sensitivity by stratifying individuals based on the presence of antibiotic resistance genes (ARGs) or opportunistic pathogens (OPs) in the baseline fecal microbiota. Quantitative Polymerase Chain Reaction (qPCR) was used to detect ARGs and OPs in DNA isolated from fecal samples of 56 males with overweight/obesity (Body Mass Index: 25-35 kg/m2) and impaired glucose metabolism (fasting plasma glucose ≥5.6 mmol/L and/or 2-hour glucose 7.8-11.1 mmol/L). A two-step hyperinsulinemic-euglycemic clamp was performed to determine tissue-specific insulin sensitivity. Abdominal subcutaneous adipose tissue (AT) gene expression was assessed using Affymetrix microarray. Gut microbial composition was determined using the Human Intestinal Tract Chip (HITChip) microarray. At baseline, the vancomycin resistance gene vanB was present in 60% of our population. In individuals that were vanB-negative at baseline, AT insulin sensitivity (insulin-mediated suppression of plasma free fatty acids) improved during vancomycin use, while it decreased among vanB-positive individuals (% change post versus baseline: 14.1 ± 5.6 vs. -6.7 ± 7.5% (p = .042)). The vancomycin-induced increase in AT insulin sensitivity was accompanied by downregulation of inflammatory pathways and enrichment of extracellular matrix remodeling pathways in AT. In the vanB-positive group, well-known vanB-carrying bacteria, Enterococcus and Streptococcus, expanded in the gut microbiome. In conclusion, microbiome composition and adipose tissue biology were differentially affected by vancomycin treatment based on fecal vanB carriage.
Keywords: Antibiotic resistance; adipose tissue; gut microbiome; insulin sensitivity; metabolic health; obesity; vanB; vancomycin.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
-
- The review on antimicrobial resistance cbJON . Antimicrobial resistance: tackling a crisis for the health and wealth of nations. 2014. https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Ta....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
