cytoNet: Spatiotemporal network analysis of cell communities
- PMID: 35696439
- PMCID: PMC9191702
- DOI: 10.1371/journal.pcbi.1009846
cytoNet: Spatiotemporal network analysis of cell communities
Erratum in
-
Correction: cytoNet: Spatiotemporal network analysis of cell communities.PLoS Comput Biol. 2022 Nov 8;18(11):e1010644. doi: 10.1371/journal.pcbi.1010644. eCollection 2022 Nov. PLoS Comput Biol. 2022. PMID: 36346791 Free PMC article.
-
Correction: cytoNet: Spatiotemporal network analysis of cell communities.PLoS Comput Biol. 2025 Jun 10;21(6):e1013176. doi: 10.1371/journal.pcbi.1013176. eCollection 2025 Jun. PLoS Comput Biol. 2025. PMID: 40493575 Free PMC article.
Abstract
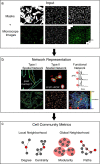
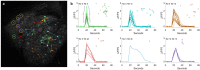
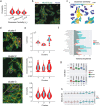
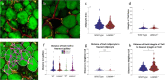
We introduce cytoNet, a cloud-based tool to characterize cell populations from microscopy images. cytoNet quantifies spatial topology and functional relationships in cell communities using principles of network science. Capturing multicellular dynamics through graph features, cytoNet also evaluates the effect of cell-cell interactions on individual cell phenotypes. We demonstrate cytoNet's capabilities in four case studies: 1) characterizing the temporal dynamics of neural progenitor cell communities during neural differentiation, 2) identifying communities of pain-sensing neurons in vivo, 3) capturing the effect of cell community on endothelial cell morphology, and 4) investigating the effect of laminin α4 on perivascular niches in adipose tissue. The analytical framework introduced here can be used to study the dynamics of complex cell communities in a quantitative manner, leading to a deeper understanding of environmental effects on cellular behavior. The versatile, cloud-based format of cytoNet makes the image analysis framework accessible to researchers across domains.
Conflict of interest statement
The authors have declared that no competing interests exist. Author David T Ryan was unable to confirm their authorship contributions. On their behalf, the corresponding author has reported their contributions to the best of their knowledge.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

