Transcriptional and functional motifs defining renal function revealed by single-nucleus RNA sequencing
- PMID: 35696569
- PMCID: PMC9231607
- DOI: 10.1073/pnas.2203179119
Transcriptional and functional motifs defining renal function revealed by single-nucleus RNA sequencing
Abstract
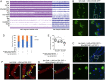
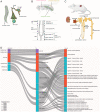
Recent advances in single-cell sequencing provide a unique opportunity to gain novel insights into the diversity, lineage, and functions of cell types constituting a tissue/organ. Here, we performed a single-nucleus study of the adult Drosophila renal system, consisting of Malpighian tubules and nephrocytes, which shares similarities with the mammalian kidney. We identified 11 distinct clusters representing renal stem cells, stellate cells, regionally specific principal cells, garland nephrocyte cells, and pericardial nephrocytes. Characterization of the transcription factors specific to each cluster identified fruitless (fru) as playing a role in stem cell regeneration and Hepatocyte nuclear factor 4 (Hnf4) in regulating glycogen and triglyceride metabolism. In addition, we identified a number of genes, including Rho guanine nucleotide exchange factor at 64C (RhoGEF64c), Frequenin 2 (Frq2), Prip, and CG1093 that are involved in regulating the unusual star shape of stellate cells. Importantly, the single-nucleus dataset allows visualization of the expression at the organ level of genes involved in ion transport and junctional permeability, providing a systems-level view of the organization and physiological roles of the tubules. Finally, a cross-species analysis allowed us to match the fly kidney cell types to mouse kidney cell types and planarian protonephridia, knowledge that will help the generation of kidney disease models. Altogether, our study provides a comprehensive resource for studying the fly kidney.
Keywords: Malpighian tubules; cross-species; kidney disease; nephrocytes; snRNA-seq.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Nielsen S., Kwon T. H., Fenton R. A., Praetorious J., “Anatomy of the kidney” in Brenner & Rector’s the Kidney, Taal M. W., et al., Eds. (Elsevier/Saunders, ed. 9, 2012), pp. 31–93.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

