Revealing the recent demographic history of Europe via haplotype sharing in the UK Biobank
- PMID: 35696575
- PMCID: PMC9233301
- DOI: 10.1073/pnas.2119281119
Revealing the recent demographic history of Europe via haplotype sharing in the UK Biobank
Abstract
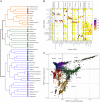
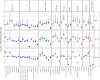
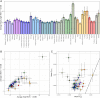
Haplotype-based analyses have recently been leveraged to interrogate the fine-scale structure in specific geographic regions, notably in Europe, although an equivalent haplotype-based understanding across the whole of Europe with these tools is lacking. Furthermore, study of identity-by-descent (IBD) sharing in a large sample of haplotypes across Europe would allow a direct comparison between different demographic histories of different regions. The UK Biobank (UKBB) is a population-scale dataset of genotype and phenotype data collected from the United Kingdom, with established sampling of worldwide ancestries. The exact content of these non-UK ancestries is largely uncharacterized, where study could highlight valuable intracontinental ancestry references with deep phenotyping within the UKBB. In this context, we sought to investigate the sample of European ancestry captured in the UKBB. We studied the haplotypes of 5,500 UKBB individuals with a European birthplace; investigated the population structure and demographic history in Europe, showing in parallel the variety of footprints of demographic history in different genetic regions around Europe; and expand knowledge of the genetic landscape of the east and southeast of Europe. Providing an updated map of European genetics, we leverage IBD-segment sharing to explore the extent of population isolation and size across the continent. In addition to building and expanding upon previous knowledge in Europe, our results show the UKBB as a source of diverse ancestries beyond Britain. These worldwide ancestries sampled in the UKBB may complement and inform researchers interested in specific communities or regions not limited to Britain.
Keywords: demographic history; haplotypes; identity by descent; population genetics.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Genome of the Netherlands Consortium, Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 46, 818–825 (2014). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

