Host protease activity classifies pneumonia etiology
- PMID: 35696579
- PMCID: PMC9231472
- DOI: 10.1073/pnas.2121778119
Host protease activity classifies pneumonia etiology
Abstract
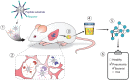
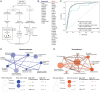
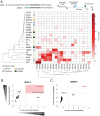
Community-acquired pneumonia (CAP) has been brought to the forefront of global health priorities due to the COVID-19 pandemic. However, classification of viral versus bacterial pneumonia etiology remains a significant clinical challenge. To this end, we have engineered a panel of activity-based nanosensors that detect the dysregulated activity of pulmonary host proteases implicated in the response to pneumonia-causing pathogens and produce a urinary readout of disease. The nanosensor targets were selected based on a human protease transcriptomic signature for pneumonia etiology generated from 33 unique publicly available study cohorts. Five mouse models of bacterial or viral CAP were developed to assess the ability of the nanosensors to produce etiology-specific urinary signatures. Machine learning algorithms were used to train diagnostic classifiers that could distinguish infected mice from healthy controls and differentiate those with bacterial versus viral pneumonia with high accuracy. This proof-of-concept diagnostic approach demonstrates a way to distinguish pneumonia etiology based solely on the host proteolytic response to infection.
Keywords: bacterial infections; diagnostics; nanoparticles; pneumonia; viral infections.
Conflict of interest statement
Competing interest statement: S.N.B. reports compensation for cofounding, consulting, and/or board membership in Glympse Bio, Satellite Bio, CEND Therapeutics, Xilio Therapeutics, Catalio Capital, Intergalactic Therapeutics, Port Therapeutics, Vertex Pharmaceuticals, Danaher, and Moderna and receives sponsored research funding from Johnson & Johnson, Revitope, and Owlstone Medical.
Figures



References
-
- Jones B. E., et al. , Summary for clinicians. Ann. Am. Thorac. Soc. 17, 133–138 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

